Clustering Linear and Nonlinear Manifolds using Generalized Principal




![Finding a basis for each subspace Polynomial Differentiation (GPCA-PDA) [CVPR’ 04] • To learn Finding a basis for each subspace Polynomial Differentiation (GPCA-PDA) [CVPR’ 04] • To learn](https://slidetodoc.com/presentation_image_h/8d7a898301b458cd5e90dd793484135b/image-15.jpg)






- Slides: 37

Clustering Linear and Nonlinear Manifolds using Generalized Principal Components Analysis René Vidal Center for Imaging Science Institute for Computational Medicine Johns Hopkins University

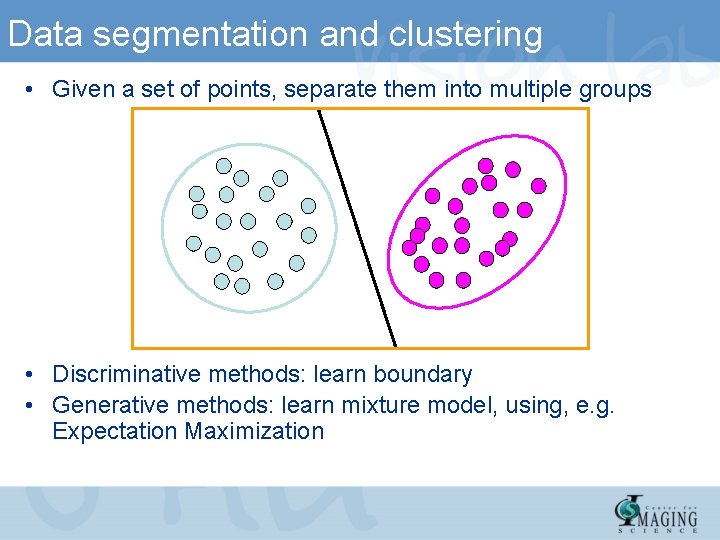
Data segmentation and clustering • Given a set of points, separate them into multiple groups • Discriminative methods: learn boundary • Generative methods: learn mixture model, using, e. g. Expectation Maximization

Dimensionality reduction and clustering • In many problems data is high-dimensional: can reduce dimensionality using, e. g. Principal Component Analysis • Image compression • Recognition – Faces (Eigenfaces) • Image segmentation – Intensity (black-white) – Texture

Clustering data on non Euclidean spaces • Clustering data on non Euclidean spaces – Mixtures of linear spaces – Mixtures of algebraic varieties – Mixtures of Lie groups • “Chicken-and-egg” problems – Given segmentation, estimate models – Given models, segment the data – Initialization? • Need to combine – Algebra/geometry, dynamics and statistics

Part I Linear Subspaces

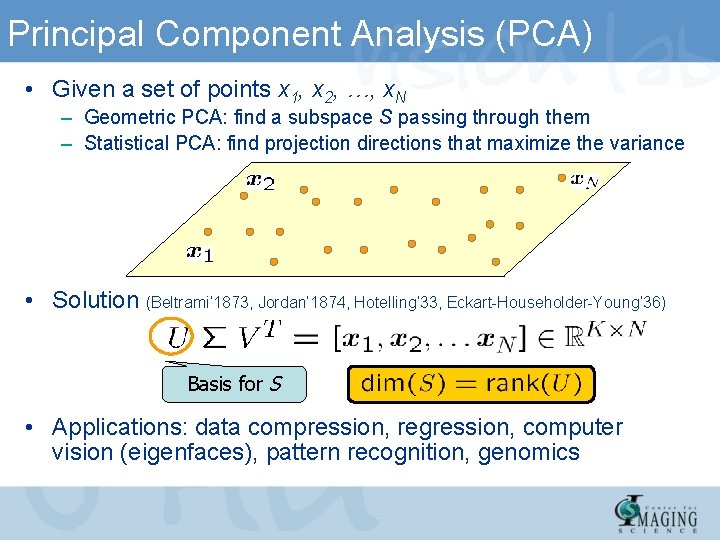
Principal Component Analysis (PCA) • Given a set of points x 1, x 2, …, x. N – Geometric PCA: find a subspace S passing through them – Statistical PCA: find projection directions that maximize the variance • Solution (Beltrami’ 1873, Jordan’ 1874, Hotelling’ 33, Eckart-Householder-Young’ 36) Basis for S • Applications: data compression, regression, computer vision (eigenfaces), pattern recognition, genomics

Generalized Principal Component Analysis • Given a set of points lying in multiple subspaces, identify – The number of subspaces and their dimensions – A basis for each subspace – The segmentation of the data points • “Chicken-and-egg” problem – Given segmentation, estimate subspaces – Given subspaces, segment the data

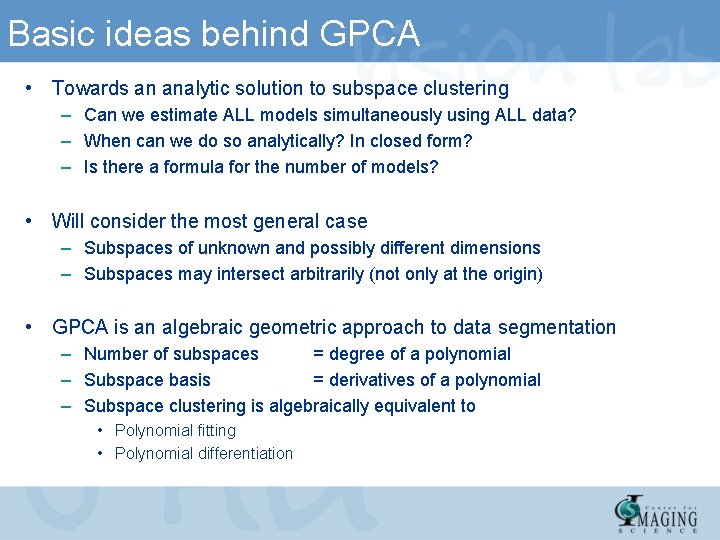
Basic ideas behind GPCA • Towards an analytic solution to subspace clustering – Can we estimate ALL models simultaneously using ALL data? – When can we do so analytically? In closed form? – Is there a formula for the number of models? • Will consider the most general case – Subspaces of unknown and possibly different dimensions – Subspaces may intersect arbitrarily (not only at the origin) • GPCA is an algebraic geometric approach to data segmentation – Number of subspaces = degree of a polynomial – Subspace basis = derivatives of a polynomial – Subspace clustering is algebraically equivalent to • Polynomial fitting • Polynomial differentiation

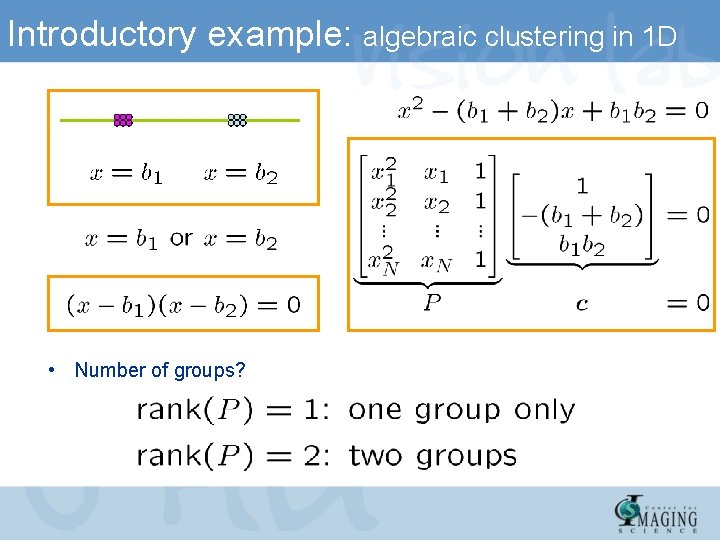
Introductory example: algebraic clustering in 1 D • Number of groups?

Introductory example: algebraic clustering in 1 D • How to compute n, c, b’s? – Number of clusters – Cluster centers – Solution is unique if – Solution is closed form if

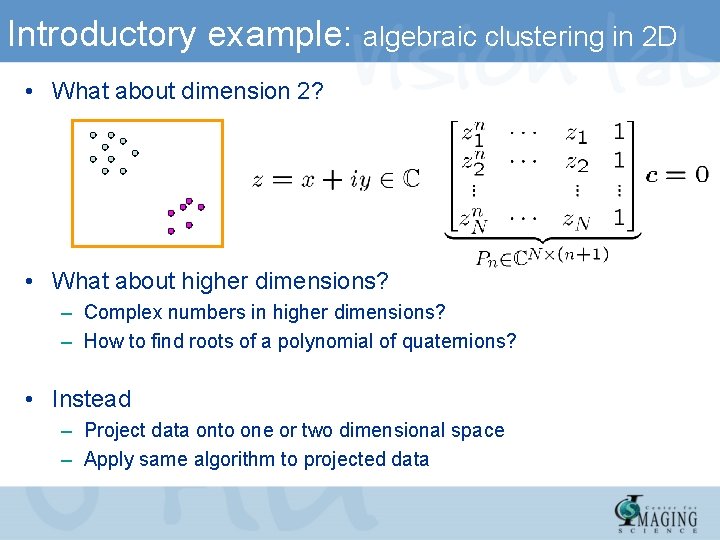
Introductory example: algebraic clustering in 2 D • What about dimension 2? • What about higher dimensions? – Complex numbers in higher dimensions? – How to find roots of a polynomial of quaternions? • Instead – Project data onto one or two dimensional space – Apply same algorithm to projected data

Representing one subspace • One plane • One line • One subspace can be represented with – Set of linear equations – Set of polynomials of degree 1

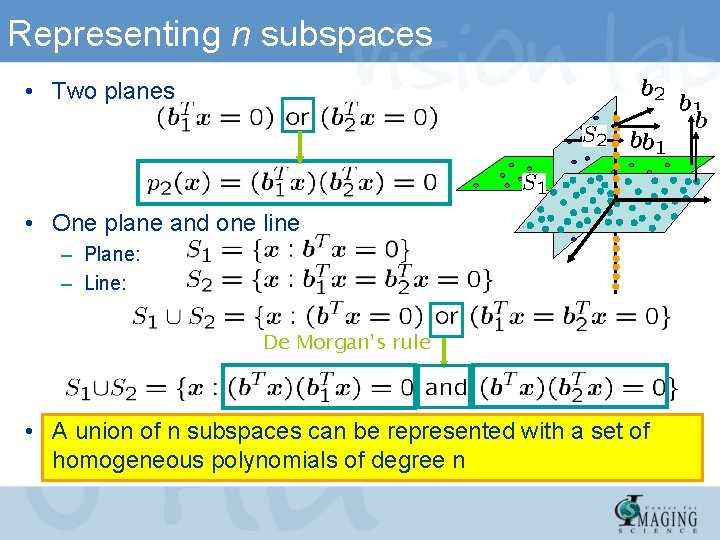
Representing n subspaces • Two planes • One plane and one line – Plane: – Line: De Morgan’s rule • A union of n subspaces can be represented with a set of homogeneous polynomials of degree n

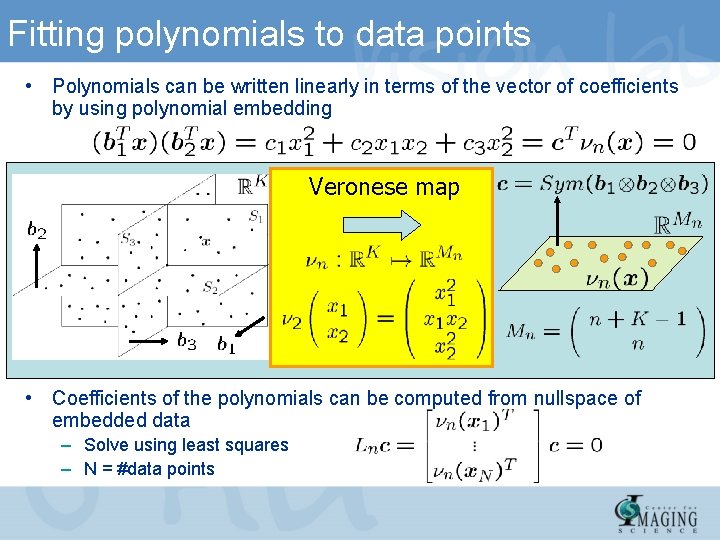
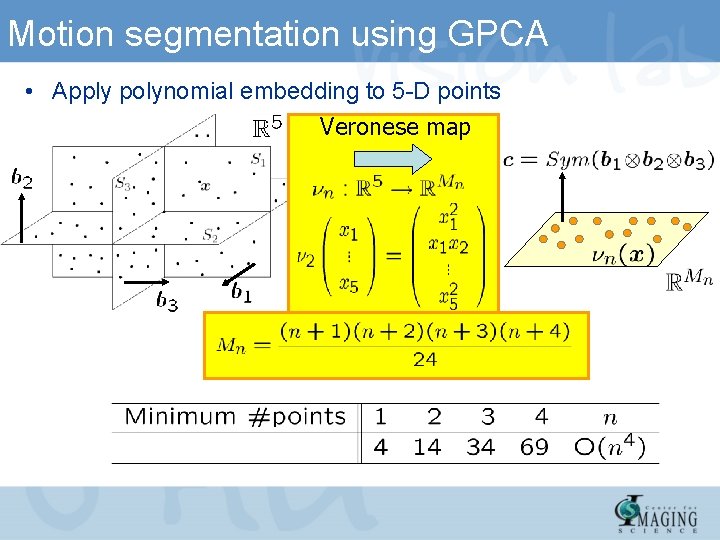
Fitting polynomials to data points • Polynomials can be written linearly in terms of the vector of coefficients by using polynomial embedding Veronese map • Coefficients of the polynomials can be computed from nullspace of embedded data – Solve using least squares – N = #data points
![Finding a basis for each subspace Polynomial Differentiation GPCAPDA CVPR 04 To learn Finding a basis for each subspace Polynomial Differentiation (GPCA-PDA) [CVPR’ 04] • To learn](https://slidetodoc.com/presentation_image_h/8d7a898301b458cd5e90dd793484135b/image-15.jpg)
Finding a basis for each subspace Polynomial Differentiation (GPCA-PDA) [CVPR’ 04] • To learn a mixture of subspaces we just need one positive example per class

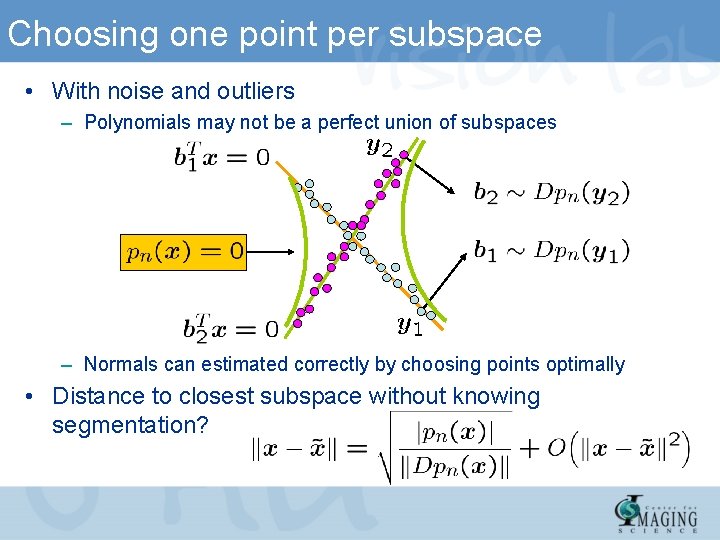
Choosing one point per subspace • With noise and outliers – Polynomials may not be a perfect union of subspaces – Normals can estimated correctly by choosing points optimally • Distance to closest subspace without knowing segmentation?

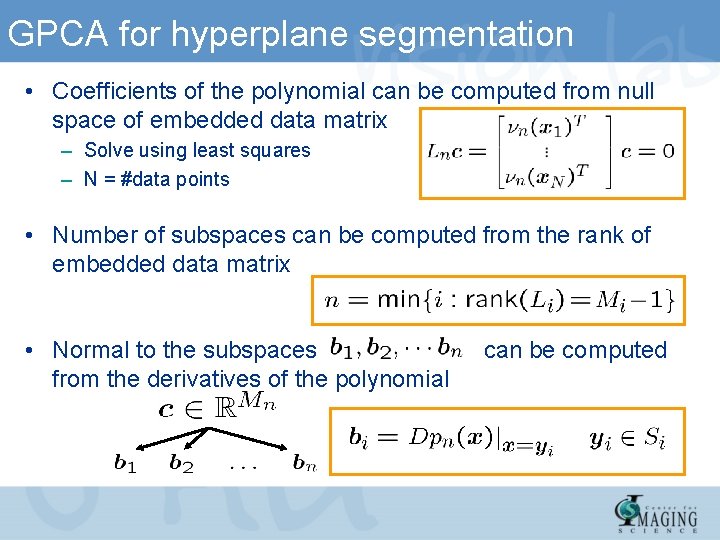
GPCA for hyperplane segmentation • Coefficients of the polynomial can be computed from null space of embedded data matrix – Solve using least squares – N = #data points • Number of subspaces can be computed from the rank of embedded data matrix • Normal to the subspaces from the derivatives of the polynomial can be computed

Motion segmentation using GPCA • Apply polynomial embedding to 5 -D points Veronese map

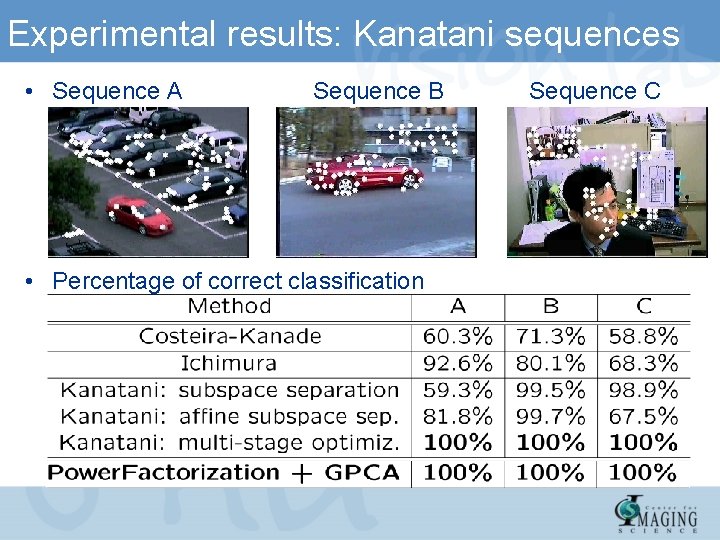
Experimental results: Kanatani sequences • Sequence A Sequence B • Percentage of correct classification Sequence C

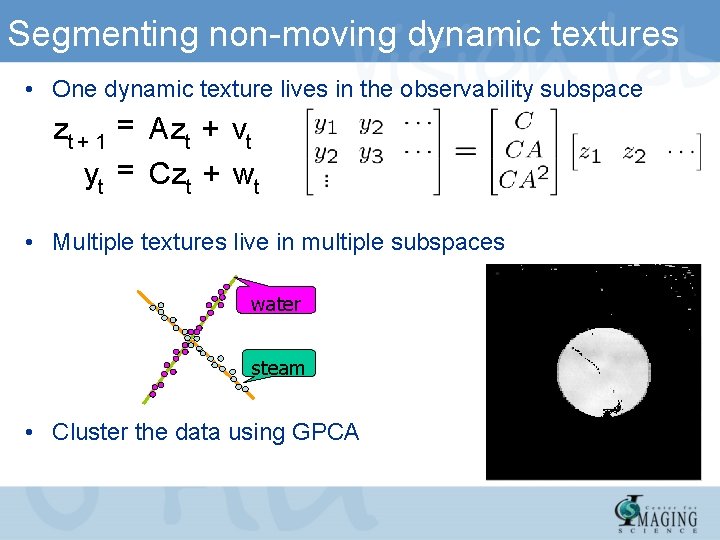
Segmenting non-moving dynamic textures • One dynamic texture lives in the observability subspace zt + 1 = Azt + vt yt = Czt + wt • Multiple textures live in multiple subspaces water steam • Cluster the data using GPCA

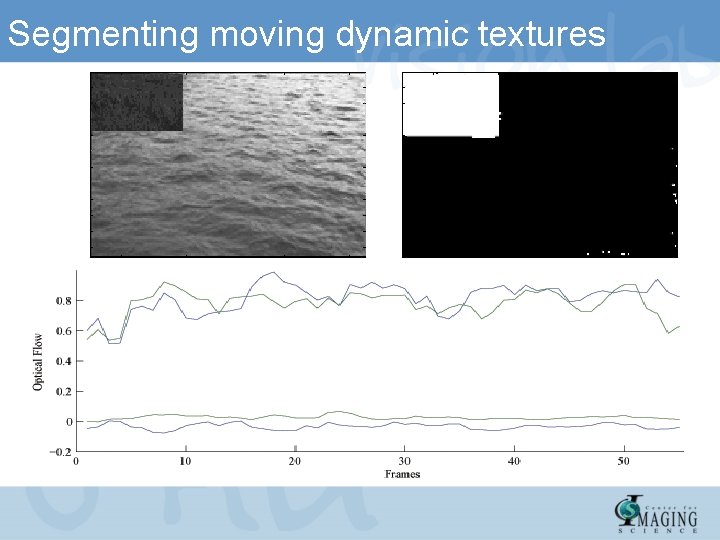
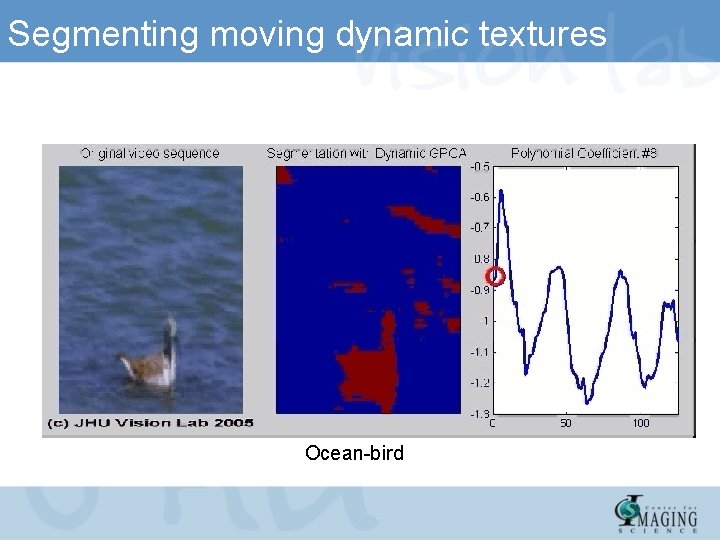
Segmenting moving dynamic textures

Segmenting moving dynamic textures Ocean-bird

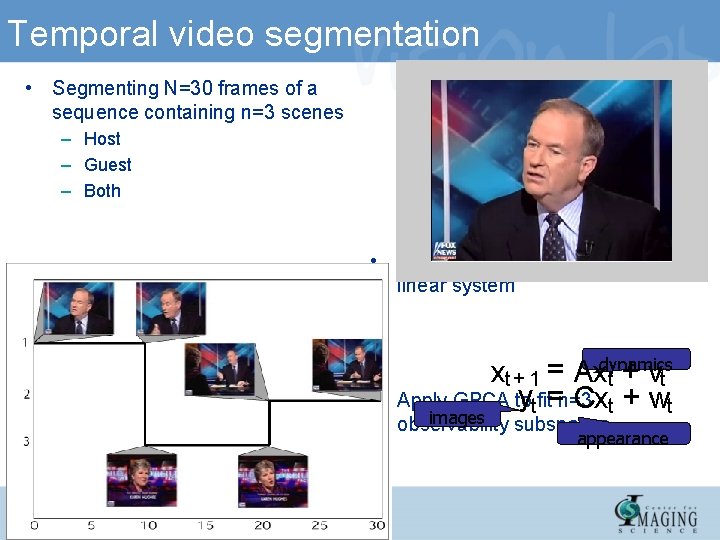
Temporal video segmentation • Segmenting N=30 frames of a sequence containing n=3 scenes – Host – Guest – Both • Image intensities are output of linear system • xt + 1 = Axdynamics t + vt ytfit=n=3 Apply GPCA to Cxt + wt images observability subspaces appearance

Temporal video segmentation • Segmenting N=60 frames of a sequence containing n=3 scenes – Burning wheel – Burnt car with people – Burning car • Image intensities are output of linear system • xt + 1 = Axdynamics t + vt ytfit=n=3 Apply GPCA to Cxt + wt images observability subspaces appearance

Part 2 Submanifolds in Riemannian Spaces

Overview of our approach • Propose a novel framework for simultaneous dimensionality reduction and clustering of data lying in different submanifolds of a Riemannian space. • Extend nonlinear dimensionality reduction techniques from one submanifold of to m submanifolds of a Riemannian space. • Show that when the different submanifolds are separated, all the points from one connected submanifold can be mapped to a single point in a lowdimensional space.

Nonlinear Dimensionality Reduction • Two kinds of dimensionality reduction: global and local techniques. • Global techniques – preserve global properties of the data lying on a submanifold – similar to PCA for a linear subspace – Eg: ISOMAP and Kernel PCA • Local techniques – preserve local properties, obtained from the small neighborhoods around the datapoints. – also retain the global properties of the data – Eg: Locally Linear Embedding (LLE), Laplacian Eigenmaps, Hessian LLE

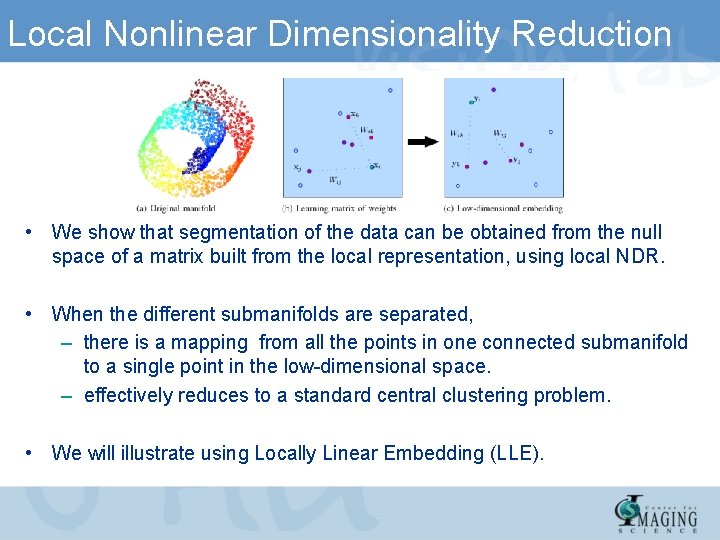
Local Nonlinear Dimensionality Reduction • We show that segmentation of the data can be obtained from the null space of a matrix built from the local representation, using local NDR. • When the different submanifolds are separated, – there is a mapping from all the points in one connected submanifold to a single point in the low-dimensional space. – effectively reduces to a standard central clustering problem. • We will illustrate using Locally Linear Embedding (LLE).

Local Nonlinear Dimensionality Reduction • Focus on local NDR methods. Each method operates in a similar manner as follows: • Step 1: Find the k-nearest neighbors of each data point according to the Euclidean distance • Step 2: Compute a matrix , that depends on the weights. represents the local geometry of the data points and incorporates reconstruction cost function in low dimension. • Step 3: Solve a sparse eigenvalue problem on matrix. The first eigenvector is the constant vector corresponding to eigenvalue 0.

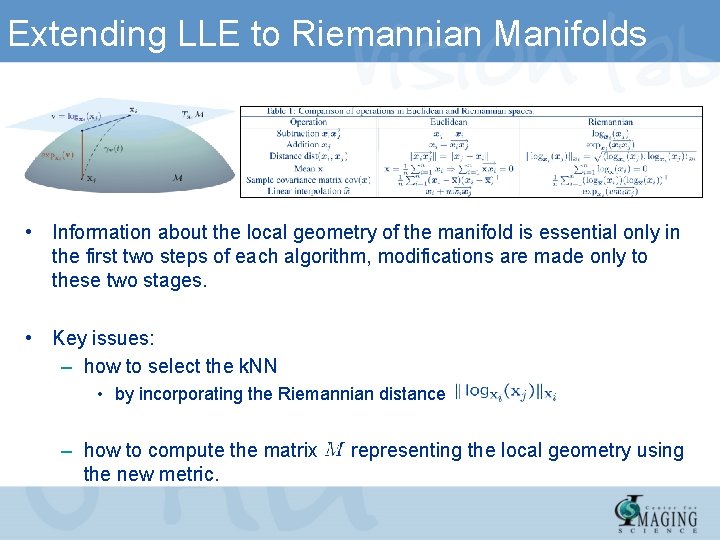
Extending LLE to Riemannian Manifolds • Information about the local geometry of the manifold is essential only in the first two steps of each algorithm, modifications are made only to these two stages. • Key issues: – how to select the k. NN • by incorporating the Riemannian distance – how to compute the matrix the new metric. representing the local geometry using

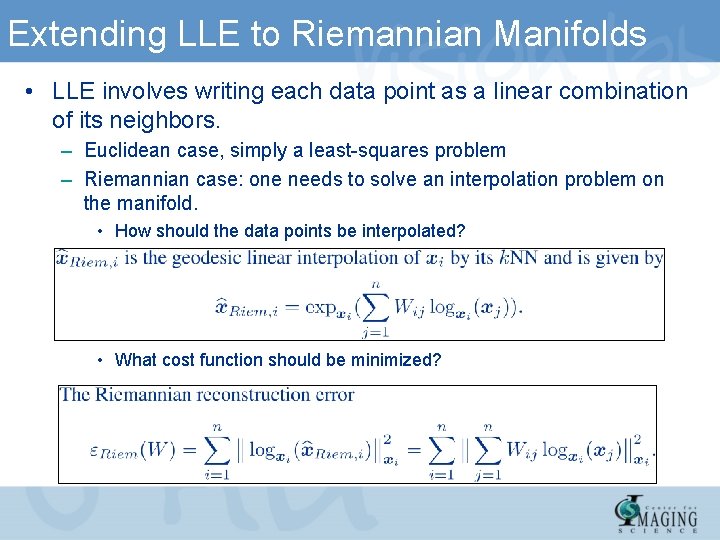
Extending LLE to Riemannian Manifolds • LLE involves writing each data point as a linear combination of its neighbors. – Euclidean case, simply a least-squares problem – Riemannian case: one needs to solve an interpolation problem on the manifold. • How should the data points be interpolated? • What cost function should be minimized?

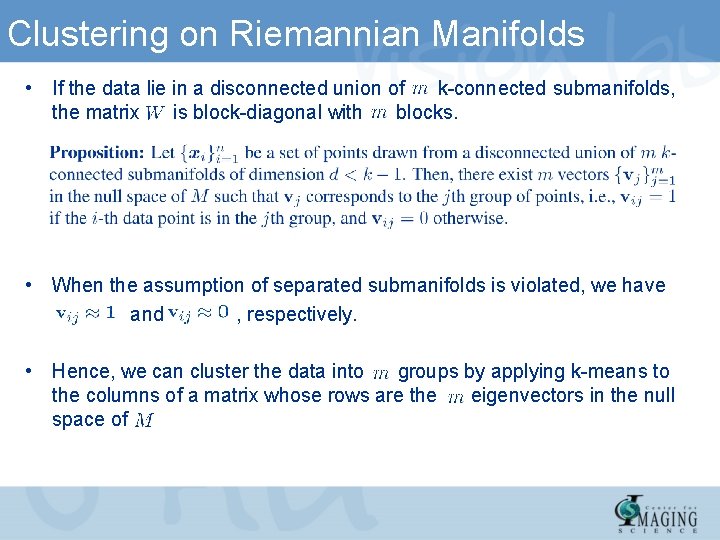
Clustering on Riemannian Manifolds • If the data lie in a disconnected union of k-connected submanifolds, the matrix is block-diagonal with blocks. • When the assumption of separated submanifolds is violated, we have and , respectively. • Hence, we can cluster the data into groups by applying k-means to the columns of a matrix whose rows are the eigenvectors in the null space of.

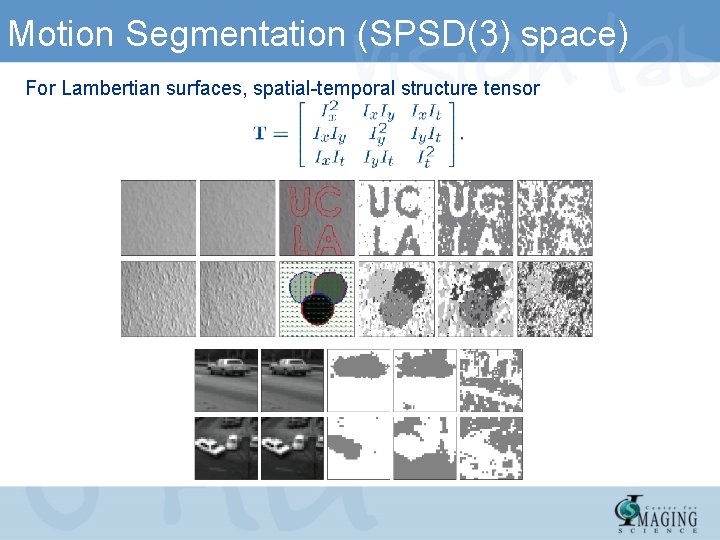
Motion Segmentation (SPSD(3) space) For Lambertian surfaces, spatial-temporal structure tensor

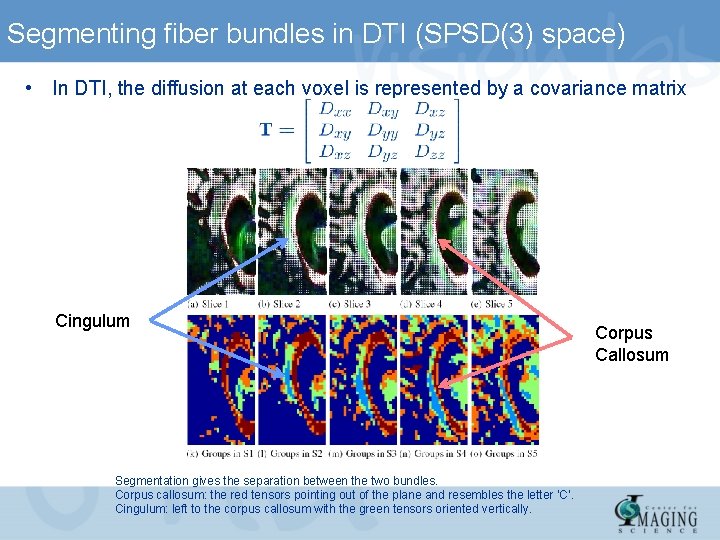
Segmenting fiber bundles in DTI (SPSD(3) space) • In DTI, the diffusion at each voxel is represented by a covariance matrix Cingulum Segmentation gives the separation between the two bundles. Corpus callosum: the red tensors pointing out of the plane and resembles the letter ‘C’. Cingulum: left to the corpus callosum with the green tensors oriented vertically. Corpus Callosum

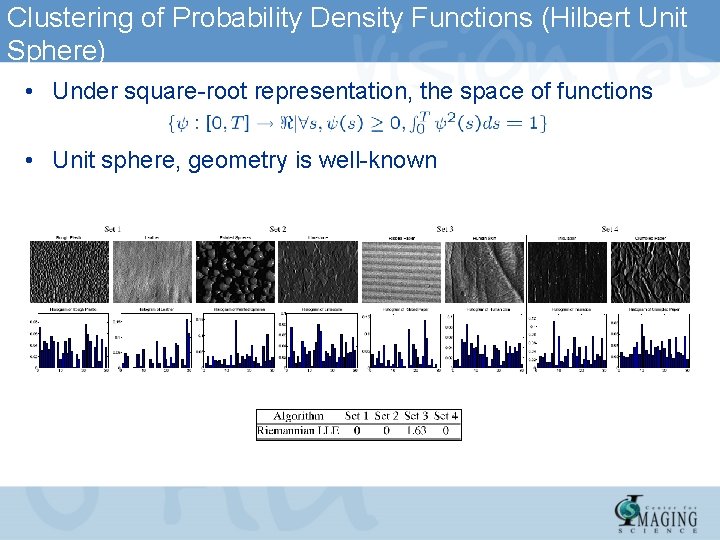
Clustering of Probability Density Functions (Hilbert Unit Sphere) • Under square-root representation, the space of functions • Unit sphere, geometry is well-known

Summary and Future Work

For more information, Vision, Dynamics and Learning Lab @ Johns Hopkins University Thank You!