Clustering Clustering Preliminaries Log 2 transformation Row centering


- Slides: 30

Clustering

Clustering Preliminaries • Log 2 transformation • Row centering and normalization • Filtering

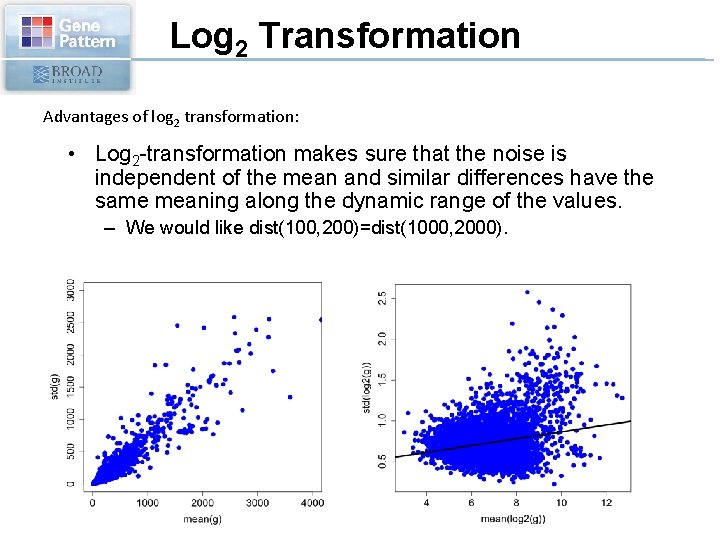
Log 2 Transformation Advantages of log 2 transformation: • Log 2 -transformation makes sure that the noise is independent of the mean and similar differences have the same meaning along the dynamic range of the values. – We would like dist(100, 200)=dist(1000, 2000).

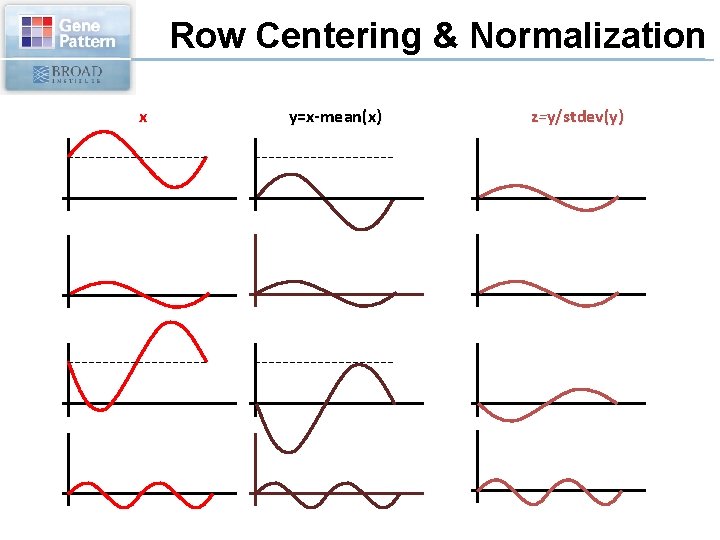
Row Centering & Normalization x y=x-mean(x) z=y/stdev(y)

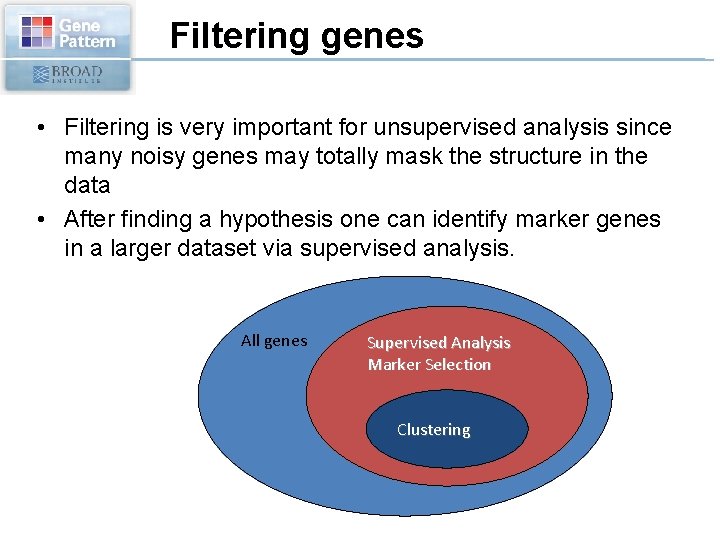
Filtering genes • Filtering is very important for unsupervised analysis since many noisy genes may totally mask the structure in the data • After finding a hypothesis one can identify marker genes in a larger dataset via supervised analysis. All genes Supervised Analysis Marker Selection Clustering

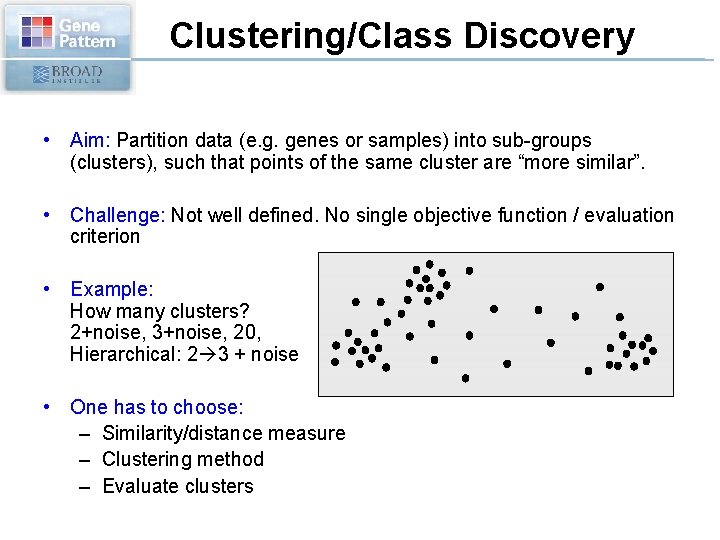
Clustering/Class Discovery • Aim: Partition data (e. g. genes or samples) into sub-groups (clusters), such that points of the same cluster are “more similar”. • Challenge: Not well defined. No single objective function / evaluation criterion • Example: How many clusters? 2+noise, 3+noise, 20, Hierarchical: 2 3 + noise • One has to choose: – Similarity/distance measure – Clustering method – Evaluate clusters

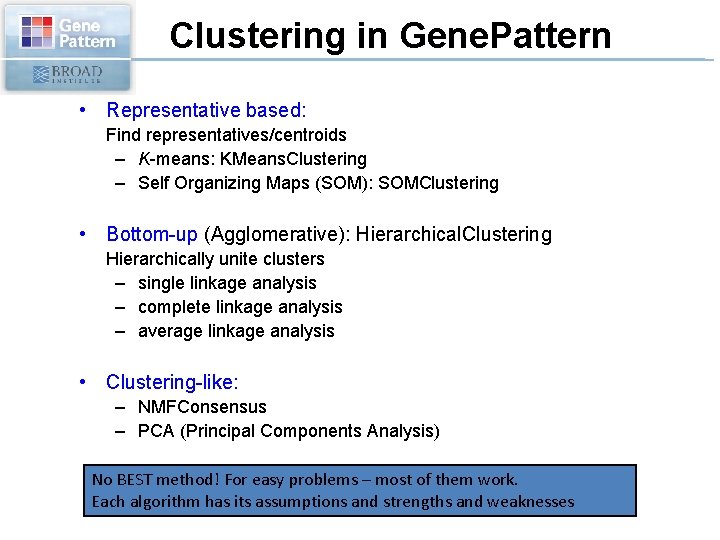
Clustering in Gene. Pattern • Representative based: Find representatives/centroids – K-means: KMeans. Clustering – Self Organizing Maps (SOM): SOMClustering • Bottom-up (Agglomerative): Hierarchical. Clustering Hierarchically unite clusters – single linkage analysis – complete linkage analysis – average linkage analysis • Clustering-like: – NMFConsensus – PCA (Principal Components Analysis) No BEST method! For easy problems – most of them work. Each algorithm has its assumptions and strengths and weaknesses

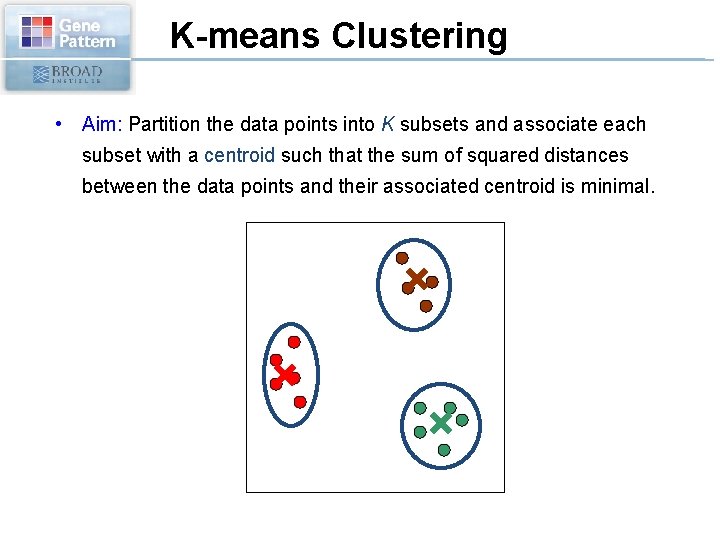
K-means Clustering • Aim: Partition the data points into K subsets and associate each subset with a centroid such that the sum of squared distances between the data points and their associated centroid is minimal.

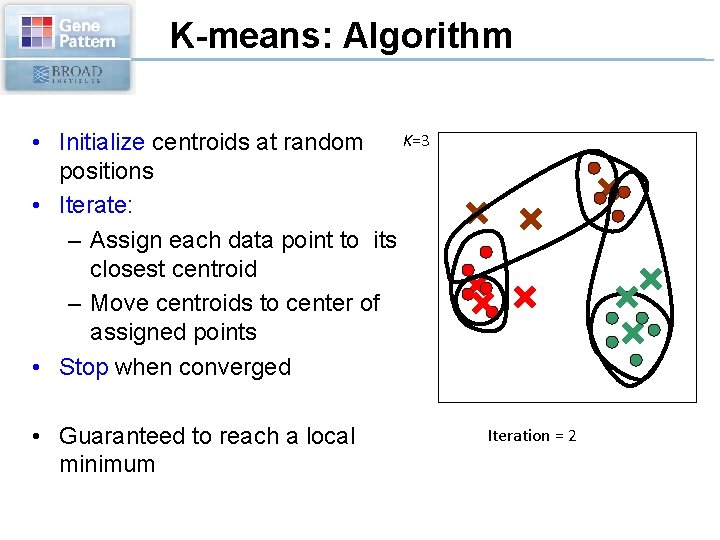
K-means: Algorithm K=3 • Initialize centroids at random positions • Iterate: – Assign each data point to its closest centroid – Move centroids to center of assigned points • Stop when converged • Guaranteed to reach a local minimum 0 Iteration = 2 1

K-means: Summary • Result depends on initial centroids’ position • Fast algorithm: needs to compute distances from data points to centroids • Must preset number of clusters. • Fails for non-spherical distributions

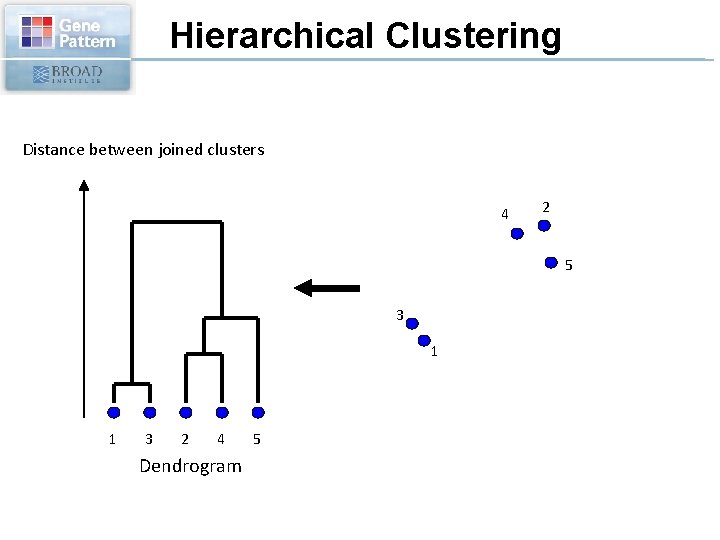
Hierarchical Clustering Distance between joined clusters 4 2 5 3 1 1 3 2 4 Dendrogram 5

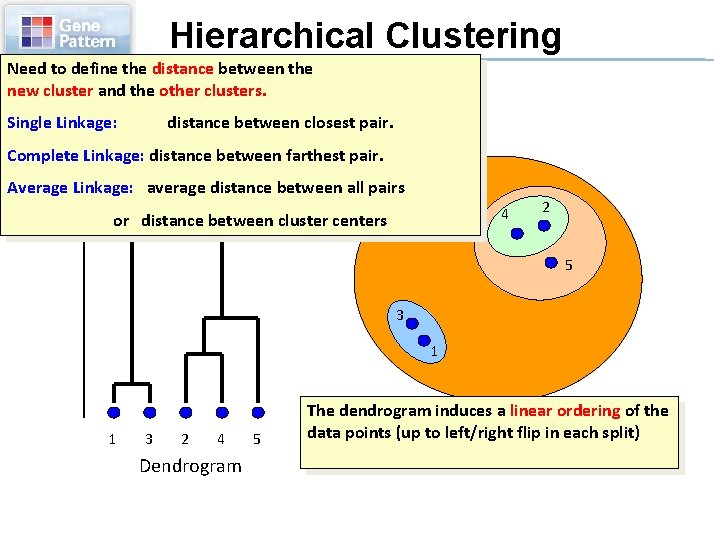
Hierarchical Clustering Need to define the distance between the new cluster and the other clusters. Single Linkage: distance between closest pair. Distance. Linkage: betweendistance joined clusters Complete between farthest pair. Average Linkage: average distance between all pairs 4 or distance between cluster centers 2 5 3 1 1 3 2 4 Dendrogram 5 The dendrogram induces a linear ordering of the data points (up to left/right flip in each split)

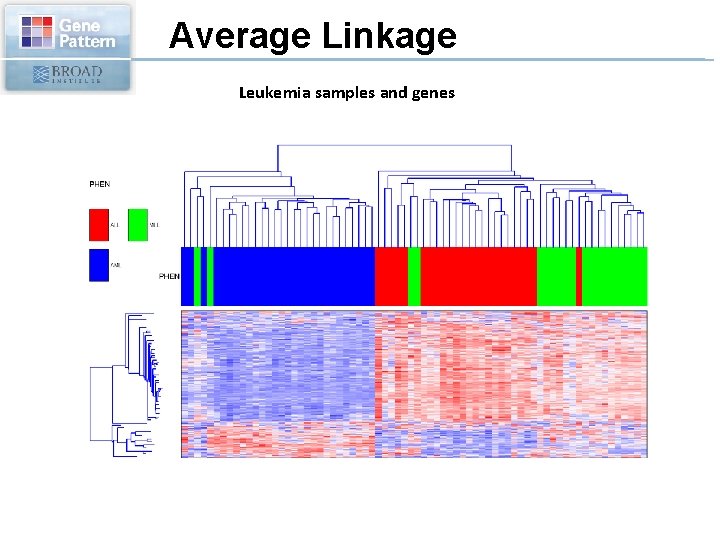
Average Linkage Leukemia samples and genes

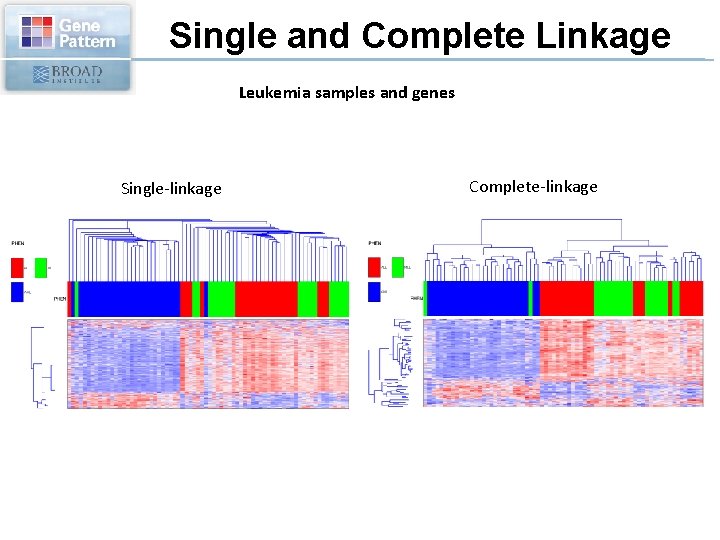
Single and Complete Linkage Leukemia samples and genes Single-linkage Complete-linkage

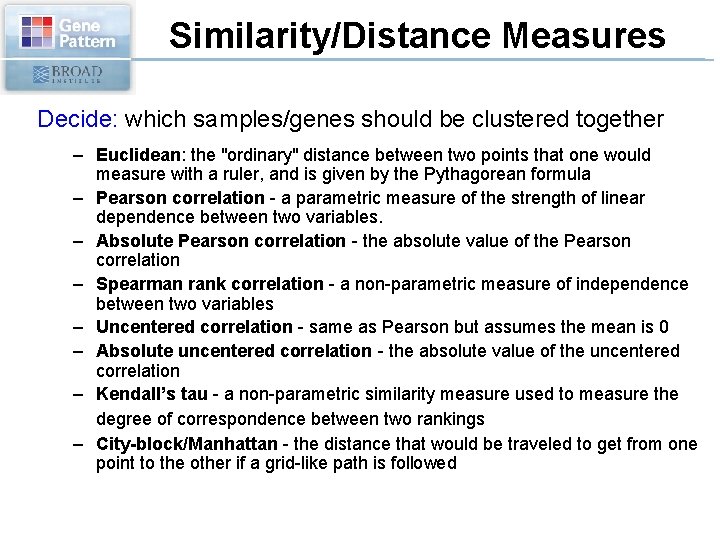
Similarity/Distance Measures Decide: which samples/genes should be clustered together – Euclidean: the "ordinary" distance between two points that one would measure with a ruler, and is given by the Pythagorean formula – Pearson correlation - a parametric measure of the strength of linear dependence between two variables. – Absolute Pearson correlation - the absolute value of the Pearson correlation – Spearman rank correlation - a non-parametric measure of independence between two variables – Uncentered correlation - same as Pearson but assumes the mean is 0 – Absolute uncentered correlation - the absolute value of the uncentered correlation – Kendall’s tau - a non-parametric similarity measure used to measure the degree of correspondence between two rankings – City-block/Manhattan - the distance that would be traveled to get from one point to the other if a grid-like path is followed

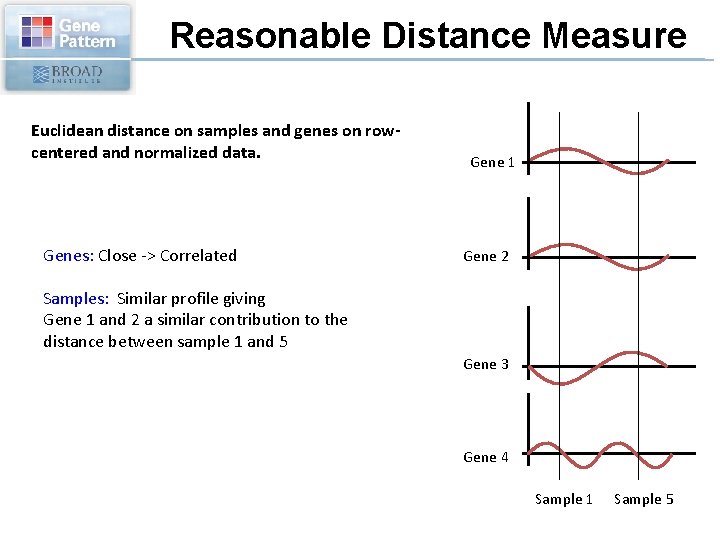
Reasonable Distance Measure Euclidean distance on samples and genes on rowcentered and normalized data. Genes: Close -> Correlated Gene 1 Gene 2 Samples: Similar profile giving Gene 1 and 2 a similar contribution to the distance between sample 1 and 5 Gene 3 Gene 4 Sample 1 Sample 5

Pitfalls in Clustering • Elongated clusters • Filament • Clusters of different sizes

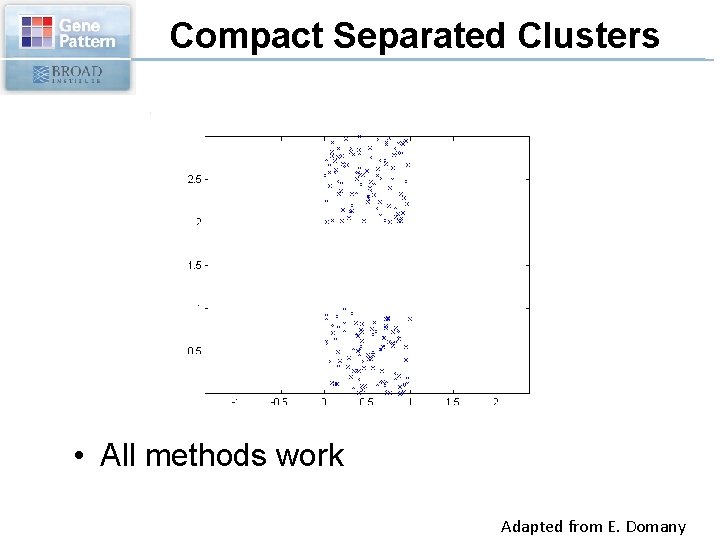
Compact Separated Clusters • All methods work Adapted from E. Domany

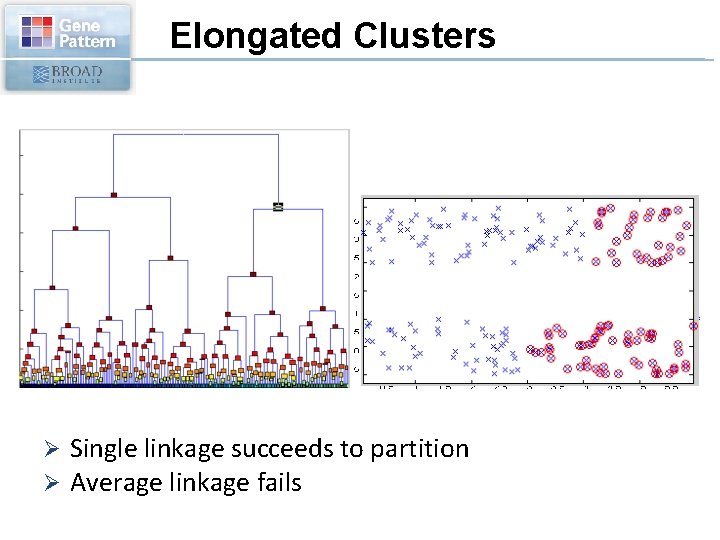
Elongated Clusters Ø Ø Single linkage succeeds to partition Average linkage fails

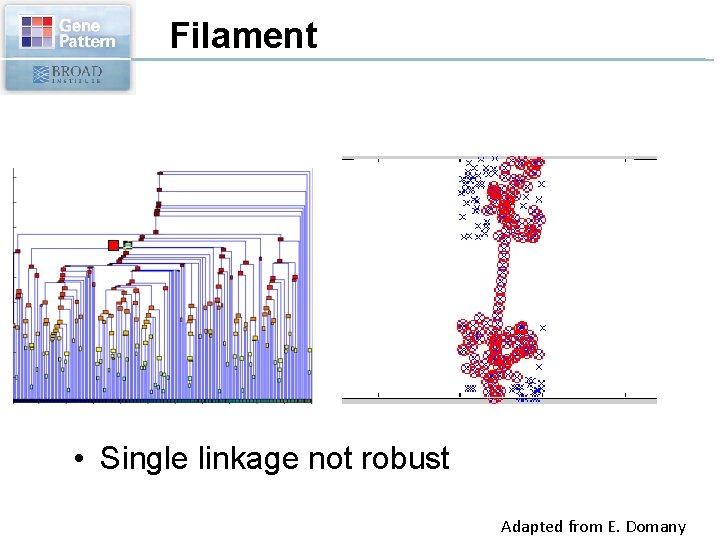
Filament • Single linkage not robust Adapted from E. Domany

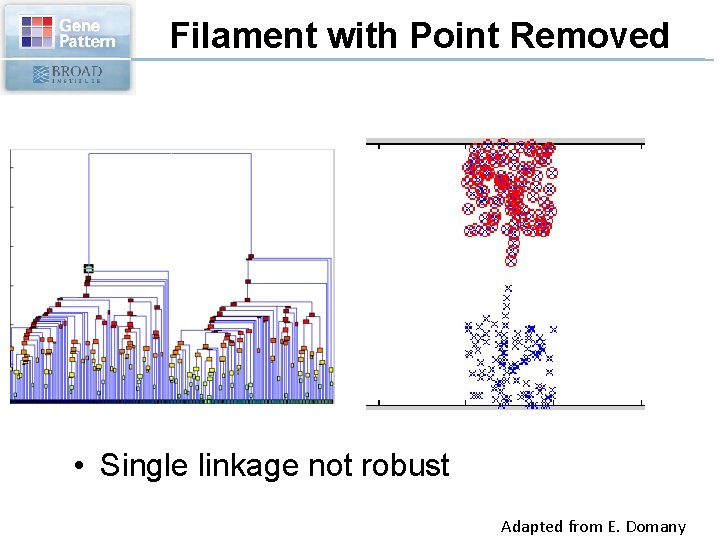
Filament with Point Removed • Single linkage not robust Adapted from E. Domany

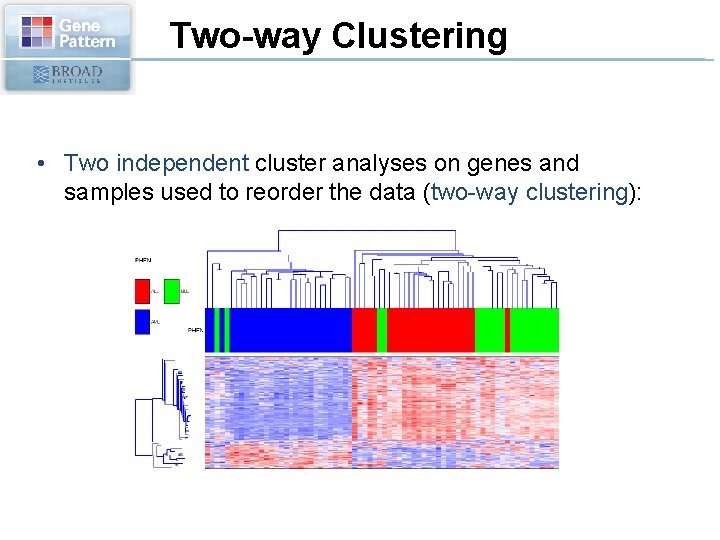
Two-way Clustering • Two independent cluster analyses on genes and samples used to reorder the data (two-way clustering):

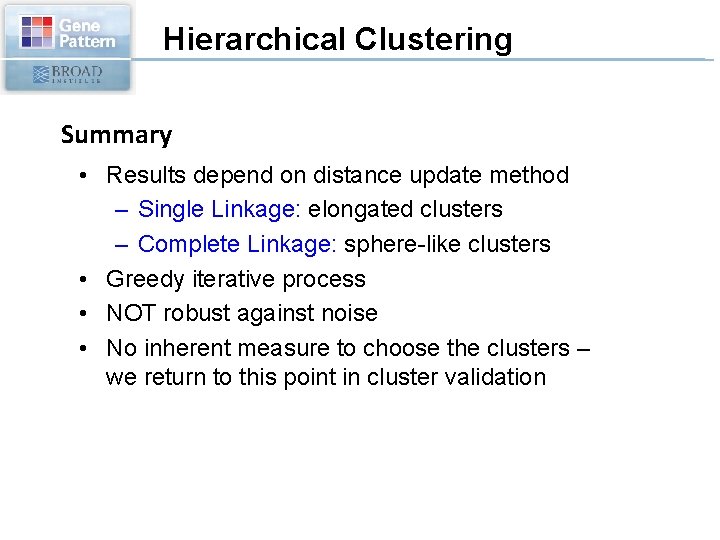
Hierarchical Clustering Summary • Results depend on distance update method – Single Linkage: elongated clusters – Complete Linkage: sphere-like clusters • Greedy iterative process • NOT robust against noise • No inherent measure to choose the clusters – we return to this point in cluster validation

Clustering Protocol

Validating Number of Clusters How do we know how many real clusters exist in the dataset?

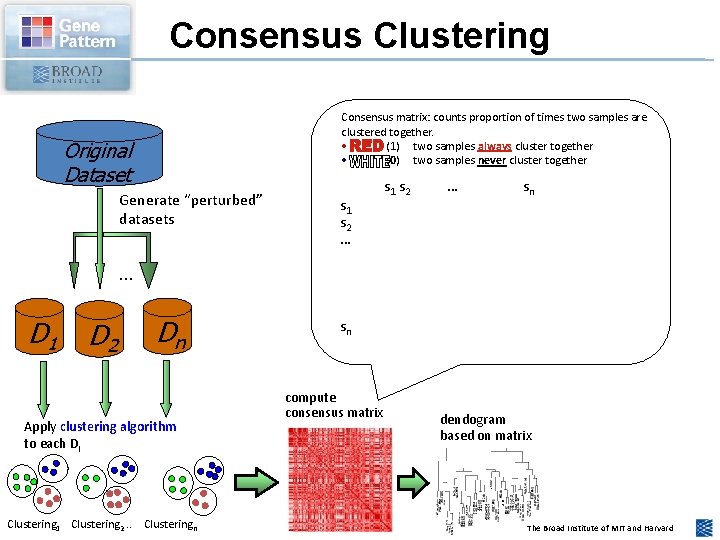
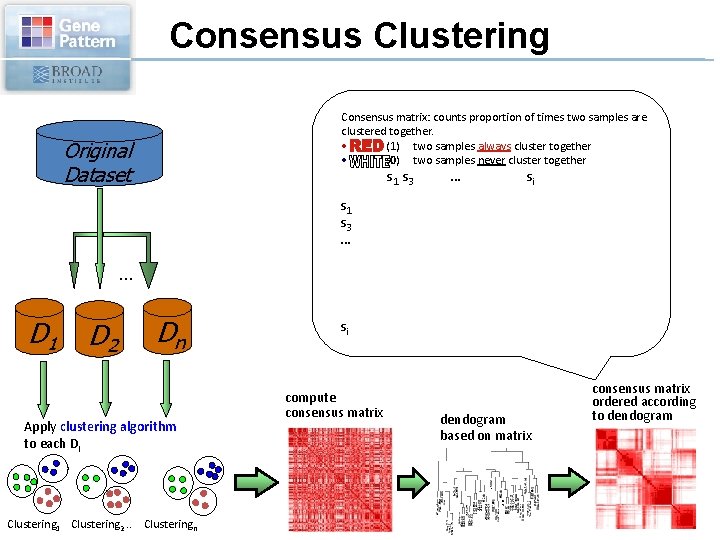
Consensus Clustering Consensus matrix: counts proportion of times two samples are clustered together. • (1) two samples always cluster together • (0) two samples never cluster together Original Dataset Generate “perturbed” datasets s 1 s 2 … sn . . . D 1 D 2 Dn Apply clustering algorithm to each Di Clustering 1 Clustering 2. . Clusteringn sn compute consensus matrix dendogram based on matrix The Broad Institute of MIT and Harvard

Consensus Clustering Consensus matrix: counts proportion of times two samples are clustered together. • (1) two samples always cluster together • (0) two samples never cluster together Original Dataset s 1 s 3 … . . . D 1 D 2 … si C 1 C 2 Dn Apply clustering algorithm to each Di Clustering 1 Clustering 2. . Clusteringn si compute consensus matrix C 3 dendogram based on matrix consensus matrix ordered according to dendogram

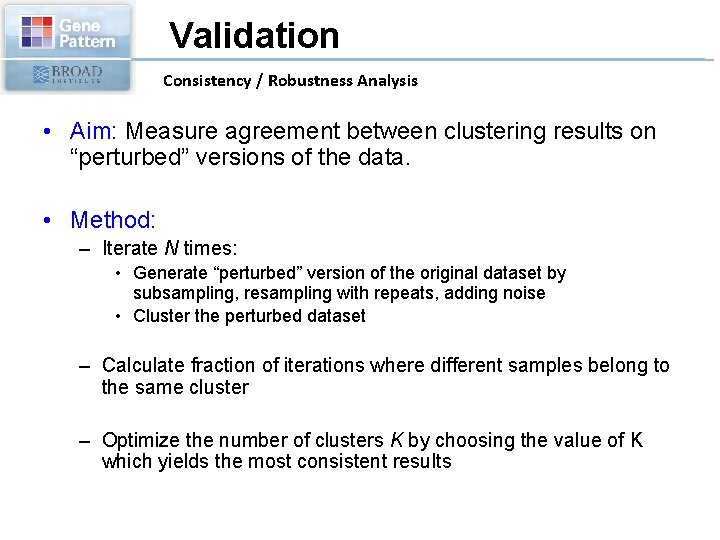
Validation Consistency / Robustness Analysis • Aim: Measure agreement between clustering results on “perturbed” versions of the data. • Method: – Iterate N times: • Generate “perturbed” version of the original dataset by subsampling, resampling with repeats, adding noise • Cluster the perturbed dataset – Calculate fraction of iterations where different samples belong to the same cluster – Optimize the number of clusters K by choosing the value of K which yields the most consistent results

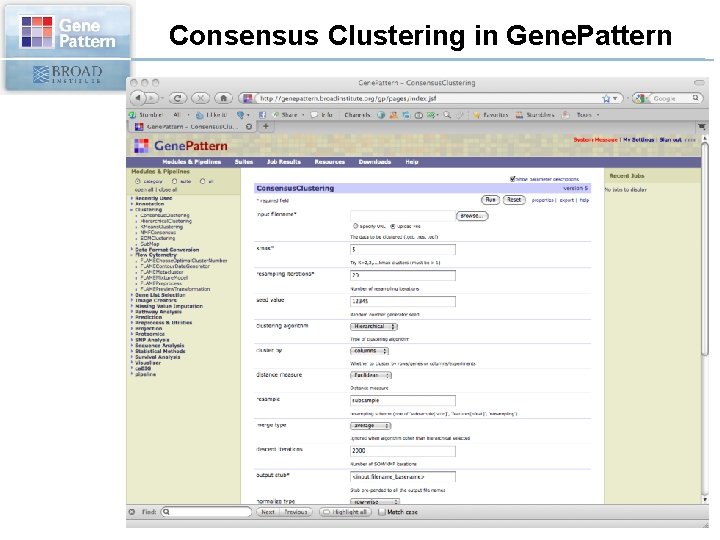
Consensus Clustering in Gene. Pattern

Clustering Cookbook • Reduce number of genes by variation filtering – Use stricter parameters than for comparative marker selection • Choose a method for cluster discovery (e. g. hierarchical clustering) • Select a number of clusters – Check for sensitivity of clusters against filtering and clustering parameters – Validate on independent data sets – Internally test robustness of clusters with consensus clustering