Clustering Analysis l l l Overview Kmeans Agglomerative


![What is Cluster Analysis? l [Chapter 8. 1. 1 , page 490] Finding groups What is Cluster Analysis? l [Chapter 8. 1. 1 , page 490] Finding groups](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-3.jpg)
![Applications of Cluster Analysis [Chapter 8. 1. 1] l Understanding – Group related documents Applications of Cluster Analysis [Chapter 8. 1. 1] l Understanding – Group related documents](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-4.jpg)
![What is not Cluster Analysis? l [Chapter 8. 1. 1 ] Supervised classification – What is not Cluster Analysis? l [Chapter 8. 1. 1 ] Supervised classification –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-5.jpg)
![Notion of a Cluster can be Ambiguous [Chapter 8. 1. 1] 2010/5/7 How many Notion of a Cluster can be Ambiguous [Chapter 8. 1. 1] 2010/5/7 How many](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-6.jpg)
![Types of Clusterings [Chapter 8. 1. 2 , page 491] l A clustering is Types of Clusterings [Chapter 8. 1. 2 , page 491] l A clustering is](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-7.jpg)
![Partitional Clustering Original Points 2010/5/7 [Chapter 8. 1. 2 , page 491] A Partitional Partitional Clustering Original Points 2010/5/7 [Chapter 8. 1. 2 , page 491] A Partitional](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-8.jpg)
![Hierarchical Clustering 2010/5/7 [Chapter 8. 1. 2 , page 492] Traditional Hierarchical Clustering Traditional Hierarchical Clustering 2010/5/7 [Chapter 8. 1. 2 , page 492] Traditional Hierarchical Clustering Traditional](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-9.jpg)
![Other Distinctions Between Sets of Clusters [Chapter 8. 1. 2 , page 492] l Other Distinctions Between Sets of Clusters [Chapter 8. 1. 2 , page 492] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-10.jpg)
![Types of Clusters: Well-Separated [Chapter 8. 1. 3 , page 493] l Well-Separated Clusters: Types of Clusters: Well-Separated [Chapter 8. 1. 3 , page 493] l Well-Separated Clusters:](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-12.jpg)
![Types of Clusters: Center-Based [Chapter 8. 1. 3 , page 494] l Center-based – Types of Clusters: Center-Based [Chapter 8. 1. 3 , page 494] l Center-based –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-13.jpg)
![Types of Clusters: Contiguity-Based [Chapter 8. 1. 3 , page 494] l Contiguous Cluster Types of Clusters: Contiguity-Based [Chapter 8. 1. 3 , page 494] l Contiguous Cluster](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-14.jpg)
![Types of Clusters: Density-Based [Chapter 8. 1. 3 , page 494] l Density-based – Types of Clusters: Density-Based [Chapter 8. 1. 3 , page 494] l Density-based –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-15.jpg)
![Types of Clusters: Conceptual Clusters [Chapter 8. 1. 3 , page 495] l Shared Types of Clusters: Conceptual Clusters [Chapter 8. 1. 3 , page 495] l Shared](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-16.jpg)
![Types of Clusters: Objective Function [optional] l Clusters Defined by an Objective Function – Types of Clusters: Objective Function [optional] l Clusters Defined by an Objective Function –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-17.jpg)
![Types of Clusters: Objective Function … [optional] l Map the clustering problem to a Types of Clusters: Objective Function … [optional] l Map the clustering problem to a](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-18.jpg)
![Characteristics of the Input Data Are Important [optional] l l Size Scale – Normalization Characteristics of the Input Data Are Important [optional] l l Size Scale – Normalization](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-19.jpg)
![K-means Clustering [Chapter 8. 2. 1 , page 497] l Partitional clustering approach l K-means Clustering [Chapter 8. 2. 1 , page 497] l Partitional clustering approach l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-21.jpg)
![K-means Clustering – Details l [Chapter 8. 2. 1] Initial centroids are often chosen K-means Clustering – Details l [Chapter 8. 2. 1] Initial centroids are often chosen](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-22.jpg)
![Two different K-means Clusterings [Chapter 8. 2. 1] Original Points Optimal Clustering 2010/5/7 Sub-optimal Two different K-means Clusterings [Chapter 8. 2. 1] Original Points Optimal Clustering 2010/5/7 Sub-optimal](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-23.jpg)
![Importance of Choosing Initial Centroids [optional] 2010/5/7 25 Importance of Choosing Initial Centroids [optional] 2010/5/7 25](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-24.jpg)
![Importance of Choosing Initial Centroids [optional] 2010/5/7 26 Importance of Choosing Initial Centroids [optional] 2010/5/7 26](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-25.jpg)
![Evaluating K-means Clusters [Chapter 8. 2. 1 , page 499] l Most common measure Evaluating K-means Clusters [Chapter 8. 2. 1 , page 499] l Most common measure](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-27.jpg)
![Importance of Choosing Initial Centroids … [Chapter 8. 2. 1 , page 501] 2010/5/7 Importance of Choosing Initial Centroids … [Chapter 8. 2. 1 , page 501] 2010/5/7](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-28.jpg)
![Importance of Choosing Initial Centroids … [Chapter 8. 2. 1 , page 501] 2010/5/7 Importance of Choosing Initial Centroids … [Chapter 8. 2. 1 , page 501] 2010/5/7](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-29.jpg)
![Problems with Selecting Initial Points [Chapter 8. 2. 1 , page 503] l 2010/5/7 Problems with Selecting Initial Points [Chapter 8. 2. 1 , page 503] l 2010/5/7](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-31.jpg)
![10 Clusters Example [optional] Starting with two initial centroids in one cluster of each 10 Clusters Example [optional] Starting with two initial centroids in one cluster of each](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-32.jpg)
![10 Clusters Example [Optional] Starting with two initial centroids in one cluster of each 10 Clusters Example [Optional] Starting with two initial centroids in one cluster of each](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-33.jpg)
![10 Clusters Example [optional] Starting with some pairs of clusters having three initial centroids, 10 Clusters Example [optional] Starting with some pairs of clusters having three initial centroids,](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-34.jpg)
![10 Clusters Example [optional] Starting with some pairs of clusters having three initial centroids, 10 Clusters Example [optional] Starting with some pairs of clusters having three initial centroids,](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-35.jpg)
![Solutions to Initial Centroids Problem [Chapter 8. 2. 1 , page 504] l Multiple Solutions to Initial Centroids Problem [Chapter 8. 2. 1 , page 504] l Multiple](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-36.jpg)
![Handling Empty Clusters [Chapter 8. 2. 2 , page 506] l Basic K-means algorithm Handling Empty Clusters [Chapter 8. 2. 2 , page 506] l Basic K-means algorithm](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-37.jpg)
![Updating Centers Incrementally [Chapter 8. 2. 2 , page 508] l In the basic Updating Centers Incrementally [Chapter 8. 2. 2 , page 508] l In the basic](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-38.jpg)
![Pre-processing and Post-processing [Chapter 8. 2. 2] l Pre-processing – Normalize the data – Pre-processing and Post-processing [Chapter 8. 2. 2] l Pre-processing – Normalize the data –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-39.jpg)
![Bisecting K-means l Bisecting K-means algorithm – 2010/5/7 [Chapter 8. 2. 3, page 509] Bisecting K-means l Bisecting K-means algorithm – 2010/5/7 [Chapter 8. 2. 3, page 509]](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-40.jpg)
![Bisecting K-means Example 2010/5/7 [Chapter 8. 2. 3] 42 Bisecting K-means Example 2010/5/7 [Chapter 8. 2. 3] 42](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-41.jpg)
![Limitations of K-means l [Chapter 8. 2. 4] K-means has problems when clusters are Limitations of K-means l [Chapter 8. 2. 4] K-means has problems when clusters are](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-42.jpg)
![Limitations of K-means: Differing Sizes [Chapter 8. 2. 4 , page 511] Original Points Limitations of K-means: Differing Sizes [Chapter 8. 2. 4 , page 511] Original Points](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-43.jpg)
![Limitations of K-means: Differing Density [Chapter 8. 2. 4 , page 511] Original Points Limitations of K-means: Differing Density [Chapter 8. 2. 4 , page 511] Original Points](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-44.jpg)
![Limitations of K-means: Non-globular Shapes [Chapter 8. 2. 4 , page 511] Original Points Limitations of K-means: Non-globular Shapes [Chapter 8. 2. 4 , page 511] Original Points](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-45.jpg)
![Overcoming K-means Limitations [Chapter 8. 2. 4_optional] Original Points K-means Clusters One solution is Overcoming K-means Limitations [Chapter 8. 2. 4_optional] Original Points K-means Clusters One solution is](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-46.jpg)
![Overcoming K-means Limitations[Chapter 8. 2. 4_optional] Original Points 2010/5/7 K-means Clusters 48 Overcoming K-means Limitations[Chapter 8. 2. 4_optional] Original Points 2010/5/7 K-means Clusters 48](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-47.jpg)
![Overcoming K-means Limitations [Chapter 8. 2. 4_optional] Original Points 2010/5/7 K-means Clusters 49 Overcoming K-means Limitations [Chapter 8. 2. 4_optional] Original Points 2010/5/7 K-means Clusters 49](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-48.jpg)
![Hierarchical Clustering [Chapter 8. 3 , page 515 -516] Produces a set of nested Hierarchical Clustering [Chapter 8. 3 , page 515 -516] Produces a set of nested](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-49.jpg)
![Strengths of Hierarchical Clustering [chapter 8. 3] l Do not have to assume any Strengths of Hierarchical Clustering [chapter 8. 3] l Do not have to assume any](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-50.jpg)
![Hierarchical Clustering l [Chapter 8. 3 , page 515] Two main types of hierarchical Hierarchical Clustering l [Chapter 8. 3 , page 515] Two main types of hierarchical](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-51.jpg)
![Agglomerative Clustering Algorithm [Chapter 8. 3. 1 , page 516] l More popular hierarchical Agglomerative Clustering Algorithm [Chapter 8. 3. 1 , page 516] l More popular hierarchical](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-52.jpg)
![Starting Situation l [optional] Start with clusters of individual points and a proximity matrix Starting Situation l [optional] Start with clusters of individual points and a proximity matrix](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-53.jpg)
![Intermediate Situation l [optional] After some merging steps, we have some clusters C 1 Intermediate Situation l [optional] After some merging steps, we have some clusters C 1](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-54.jpg)
![Intermediate Situation l [optional] We want to merge the two closest clusters (C 2 Intermediate Situation l [optional] We want to merge the two closest clusters (C 2](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-55.jpg)
![After Merging l [optional] The question is “How do we update the proximity matrix? After Merging l [optional] The question is “How do we update the proximity matrix?](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-56.jpg)
![How to Define Inter-Cluster Similarity [Chapter 8. 3. 1 , page 517] p 1 How to Define Inter-Cluster Similarity [Chapter 8. 3. 1 , page 517] p 1](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-57.jpg)
![How to Define Inter-Cluster Similarity[Chapter 8. 3. 1] p 1 p 2 p 3 How to Define Inter-Cluster Similarity[Chapter 8. 3. 1] p 1 p 2 p 3](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-58.jpg)
![How to Define Inter-Cluster Similarity[Chapter 8. 3. 1] p 1 p 2 p 3 How to Define Inter-Cluster Similarity[Chapter 8. 3. 1] p 1 p 2 p 3](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-59.jpg)
![How to Define Inter-Cluster Similarity [Chapter 8. 3. 1] p 1 p 2 p How to Define Inter-Cluster Similarity [Chapter 8. 3. 1] p 1 p 2 p](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-61.jpg)
![Cluster Similarity: MIN or Single Link [Chapter 8. 3. 2 , page 519] l Cluster Similarity: MIN or Single Link [Chapter 8. 3. 2 , page 519] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-62.jpg)
![Hierarchical Clustering: MIN [Chapter 8. 3. 2 , page 520] 1 3 5 2 Hierarchical Clustering: MIN [Chapter 8. 3. 2 , page 520] 1 3 5 2](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-63.jpg)
![Strength of MIN Original Points [Chapter 8. 3. 2 ] Two Clusters • Can Strength of MIN Original Points [Chapter 8. 3. 2 ] Two Clusters • Can](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-64.jpg)
![Limitations of MIN Original Points [Chapter 8. 3. 2] Two Clusters • Sensitive to Limitations of MIN Original Points [Chapter 8. 3. 2] Two Clusters • Sensitive to](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-65.jpg)
![Cluster Similarity: MAX or Complete Linkage [Chapter 8. 3. 2 , page 520] l Cluster Similarity: MAX or Complete Linkage [Chapter 8. 3. 2 , page 520] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-66.jpg)
![Hierarchical Clustering: MAX [Chapter 8. 3. 2 , page 521] 4 1 2 5 Hierarchical Clustering: MAX [Chapter 8. 3. 2 , page 521] 4 1 2 5](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-67.jpg)
![Strength of MAX Original Points [Chapter 8. 3. 2] Two Clusters • Less susceptible Strength of MAX Original Points [Chapter 8. 3. 2] Two Clusters • Less susceptible](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-68.jpg)
![Limitations of MAX Original Points [Chapter 8. 3. 2 ] Two Clusters • Tends Limitations of MAX Original Points [Chapter 8. 3. 2 ] Two Clusters • Tends](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-69.jpg)
![Cluster Similarity: Group Average [Chapter 8. 3. 2 , page 521] l Proximity of Cluster Similarity: Group Average [Chapter 8. 3. 2 , page 521] l Proximity of](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-71.jpg)
![Hierarchical Clustering: Group Average [Chapter 8. 3. 2 , page 523] 5 4 1 Hierarchical Clustering: Group Average [Chapter 8. 3. 2 , page 523] 5 4 1](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-72.jpg)
![Hierarchical Clustering: Group Average [Chapter 8. 3. 2] l Compromise between Single and Complete Hierarchical Clustering: Group Average [Chapter 8. 3. 2] l Compromise between Single and Complete](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-73.jpg)
![Cluster Similarity: Ward’s Method [Chapter 8. 3. 2 , page 523] l Similarity of Cluster Similarity: Ward’s Method [Chapter 8. 3. 2 , page 523] l Similarity of](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-74.jpg)
![Hierarchical Clustering: Time and Space requirements [Chapter 8. 3. 4] l O(N 2) space Hierarchical Clustering: Time and Space requirements [Chapter 8. 3. 4] l O(N 2) space](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-76.jpg)
![Hierarchical Clustering: Problems and Limitations [Chapter 8. 3. 5] l Once a decision is Hierarchical Clustering: Problems and Limitations [Chapter 8. 3. 5] l Once a decision is](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-77.jpg)
![DBSCAN: Core, Border, and Noise Points [Chapter 8. 4. 1 , page 528] 2010/5/7 DBSCAN: Core, Border, and Noise Points [Chapter 8. 4. 1 , page 528] 2010/5/7](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-79.jpg)
![DBSCAN Algorithm [Chapter 8. 4. 2 , page 528] Eliminate noise points l Perform DBSCAN Algorithm [Chapter 8. 4. 2 , page 528] Eliminate noise points l Perform](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-80.jpg)
![DBSCAN: Core, Border and Noise Points [Chapter 8. 4. 2 , page 530] Original DBSCAN: Core, Border and Noise Points [Chapter 8. 4. 2 , page 530] Original](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-81.jpg)
![When DBSCAN Works Well Original Points [optional] Clusters • Resistant to Noise • Can When DBSCAN Works Well Original Points [optional] Clusters • Resistant to Noise • Can](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-82.jpg)
![When DBSCAN Does NOT Work Well [optional] (Min. Pts=4, Eps=9. 75). Original Points • When DBSCAN Does NOT Work Well [optional] (Min. Pts=4, Eps=9. 75). Original Points •](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-83.jpg)
![DBSCAN: Determining EPS and Min. Pts l l l 2010/5/7 [optional] Idea is that DBSCAN: Determining EPS and Min. Pts l l l 2010/5/7 [optional] Idea is that](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-84.jpg)
![Different Aspects of Cluster Validation [Chapter 8. 5. 1 , page 533] 1. Determining Different Aspects of Cluster Validation [Chapter 8. 5. 1 , page 533] 1. Determining](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-85.jpg)
![Internal Measures: Cohesion and Separation [Chapter 8. 5. 2 , page 536] l Cluster Internal Measures: Cohesion and Separation [Chapter 8. 5. 2 , page 536] l Cluster](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-86.jpg)
![Internal Measures: Cohesion and Separation [Chapter 8. 5. 2] l Example: SSE – BSS Internal Measures: Cohesion and Separation [Chapter 8. 5. 2] l Example: SSE – BSS](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-87.jpg)
![Internal Measures: Cohesion and Separation [Chapter 8. 5. 2] l A proximity graph based Internal Measures: Cohesion and Separation [Chapter 8. 5. 2] l A proximity graph based](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-88.jpg)
![Internal Measures: Silhouette Coefficient [Chapter 8. 5. 2 , page 541] l l Silhouette Internal Measures: Silhouette Coefficient [Chapter 8. 5. 2 , page 541] l l Silhouette](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-89.jpg)
![Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 544] l Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 544] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-90.jpg)
![Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-91.jpg)
![Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-92.jpg)
![Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-93.jpg)
![Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3] DBSCAN 2010/5/7 95 Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3] DBSCAN 2010/5/7 95](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-94.jpg)
![Internal Measures: SSE [optional] l Clusters in more complicated figures aren’t well separated l Internal Measures: SSE [optional] l Clusters in more complicated figures aren’t well separated l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-95.jpg)
![Internal Measures: SSE l [optional] SSE curve for a more complicated data set SSE Internal Measures: SSE l [optional] SSE curve for a more complicated data set SSE](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-96.jpg)
![Cluster Validity l [Chapter 8. 5. 7 , page 548] For supervised classification we Cluster Validity l [Chapter 8. 5. 7 , page 548] For supervised classification we](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-98.jpg)
![Clusters found in Random Data Random Points K-means 2010/5/7 [optional] DBSCAN Complete Link 100 Clusters found in Random Data Random Points K-means 2010/5/7 [optional] DBSCAN Complete Link 100](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-99.jpg)
![Measures of Cluster Validity l [Chapter 8. 5. 7] Numerical measures that are applied Measures of Cluster Validity l [Chapter 8. 5. 7] Numerical measures that are applied](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-101.jpg)
![Measuring Cluster Validity Via Correlation [Chapter 8. 5. 7] l l Two matrices – Measuring Cluster Validity Via Correlation [Chapter 8. 5. 7] l l Two matrices –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-102.jpg)
![Measuring Cluster Validity Via Correlation [Chapter 8. 5. 7 , page 551] l Correlation Measuring Cluster Validity Via Correlation [Chapter 8. 5. 7 , page 551] l Correlation](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-103.jpg)
![Statistical Framework for SSE l [Chapter 8. 5. 8 , page 554] Example – Statistical Framework for SSE l [Chapter 8. 5. 8 , page 554] Example –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-104.jpg)
![Statistical Framework for Correlation [Chapter 8. 5. 8] l Correlation of incidence and proximity Statistical Framework for Correlation [Chapter 8. 5. 8] l Correlation of incidence and proximity](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-105.jpg)
![Final Comment on Cluster Validity [Chapter 8. 5. 8] “The validation of clustering structures Final Comment on Cluster Validity [Chapter 8. 5. 8] “The validation of clustering structures](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-106.jpg)
- Slides: 106

Clustering Analysis

l l l Overview K-means Agglomerative Hierarchical Clustering DBSCAN Cluster Evaluation 2010/5/7 2
![What is Cluster Analysis l Chapter 8 1 1 page 490 Finding groups What is Cluster Analysis? l [Chapter 8. 1. 1 , page 490] Finding groups](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-3.jpg)
What is Cluster Analysis? l [Chapter 8. 1. 1 , page 490] Finding groups of objects such that the objects in a group will be similar (or related) to one another and different from (or unrelated to) the objects in other groups Intra-cluster distances are minimized Inter-cluster distances are maximized A good clustering method will produce high quality clusters with high intra-cluster similarity low inter-cluster similarity 2010/5/7 3
![Applications of Cluster Analysis Chapter 8 1 1 l Understanding Group related documents Applications of Cluster Analysis [Chapter 8. 1. 1] l Understanding – Group related documents](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-4.jpg)
Applications of Cluster Analysis [Chapter 8. 1. 1] l Understanding – Group related documents for browsing, group genes and proteins that have similar functionality, or group stocks with similar price fluctuations l Summarization – Reduce the size of large data sets Clustering precipitation in Australia 2010/5/7 5
![What is not Cluster Analysis l Chapter 8 1 1 Supervised classification What is not Cluster Analysis? l [Chapter 8. 1. 1 ] Supervised classification –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-5.jpg)
What is not Cluster Analysis? l [Chapter 8. 1. 1 ] Supervised classification – Have class label information l Simple segmentation – Dividing students into different registration groups alphabetically, by last name l Results of a query – Groupings are a result of an external specification l Graph partitioning – Some mutual relevance and synergy, but areas are not identical 2010/5/7 6
![Notion of a Cluster can be Ambiguous Chapter 8 1 1 201057 How many Notion of a Cluster can be Ambiguous [Chapter 8. 1. 1] 2010/5/7 How many](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-6.jpg)
Notion of a Cluster can be Ambiguous [Chapter 8. 1. 1] 2010/5/7 How many clusters? Six Clusters Two Clusters Four Clusters 7
![Types of Clusterings Chapter 8 1 2 page 491 l A clustering is Types of Clusterings [Chapter 8. 1. 2 , page 491] l A clustering is](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-7.jpg)
Types of Clusterings [Chapter 8. 1. 2 , page 491] l A clustering is a set of clusters l Important distinction between hierarchical and partitional sets of clusters l Partitional Clustering – A division data objects into non-overlapping subsets (clusters) such that each data object is in exactly one subset l Hierarchical clustering – A set of nested clusters organized as a hierarchical tree 2010/5/7 8
![Partitional Clustering Original Points 201057 Chapter 8 1 2 page 491 A Partitional Partitional Clustering Original Points 2010/5/7 [Chapter 8. 1. 2 , page 491] A Partitional](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-8.jpg)
Partitional Clustering Original Points 2010/5/7 [Chapter 8. 1. 2 , page 491] A Partitional Clustering 9
![Hierarchical Clustering 201057 Chapter 8 1 2 page 492 Traditional Hierarchical Clustering Traditional Hierarchical Clustering 2010/5/7 [Chapter 8. 1. 2 , page 492] Traditional Hierarchical Clustering Traditional](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-9.jpg)
Hierarchical Clustering 2010/5/7 [Chapter 8. 1. 2 , page 492] Traditional Hierarchical Clustering Traditional Dendrogram Non-traditional Hierarchical Clustering Non-traditional Dendrogram 10
![Other Distinctions Between Sets of Clusters Chapter 8 1 2 page 492 l Other Distinctions Between Sets of Clusters [Chapter 8. 1. 2 , page 492] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-10.jpg)
Other Distinctions Between Sets of Clusters [Chapter 8. 1. 2 , page 492] l Exclusive versus non-exclusive – In non-exclusive clusterings, points may belong to multiple clusters. – Can represent multiple classes or ‘border’ points l Fuzzy versus non-fuzzy – In fuzzy clustering, a point belongs to every cluster with some weight between 0 and 1 – Weights must sum to 1 – Probabilistic clustering has similar characteristics l Partial versus complete – In some cases, we only want to cluster some of the data l Heterogeneous versus homogeneous – Cluster of widely different sizes, shapes, and densities 2010/5/7 11

Types of Clusters l Well-separated clusters l Center-based clusters l Contiguous clusters l Density-based clusters l Property or Conceptual l Described by an Objective Function 2010/5/7 [Chapter 8. 1. 3 , page 493] 12
![Types of Clusters WellSeparated Chapter 8 1 3 page 493 l WellSeparated Clusters Types of Clusters: Well-Separated [Chapter 8. 1. 3 , page 493] l Well-Separated Clusters:](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-12.jpg)
Types of Clusters: Well-Separated [Chapter 8. 1. 3 , page 493] l Well-Separated Clusters: – A cluster is a set of points such that any point in a cluster is closer (or more similar) to every other point in the cluster than to any point not in the cluster. 3 well-separated clusters 2010/5/7 13
![Types of Clusters CenterBased Chapter 8 1 3 page 494 l Centerbased Types of Clusters: Center-Based [Chapter 8. 1. 3 , page 494] l Center-based –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-13.jpg)
Types of Clusters: Center-Based [Chapter 8. 1. 3 , page 494] l Center-based – A cluster is a set of objects such that an object in a cluster is closer (more similar) to the “center” of a cluster, than to the center of any other cluster – The center of a cluster is often a centroid, the average of all the points in the cluster, or a medoid, the most “representative” point of a cluster 4 center-based clusters 2010/5/7 14
![Types of Clusters ContiguityBased Chapter 8 1 3 page 494 l Contiguous Cluster Types of Clusters: Contiguity-Based [Chapter 8. 1. 3 , page 494] l Contiguous Cluster](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-14.jpg)
Types of Clusters: Contiguity-Based [Chapter 8. 1. 3 , page 494] l Contiguous Cluster (Nearest neighbor or Transitive) – A cluster is a set of points such that a point in a cluster is closer (or more similar) to one or more other points in the cluster than to any point not in the cluster. 8 contiguous clusters 2010/5/7 15
![Types of Clusters DensityBased Chapter 8 1 3 page 494 l Densitybased Types of Clusters: Density-Based [Chapter 8. 1. 3 , page 494] l Density-based –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-15.jpg)
Types of Clusters: Density-Based [Chapter 8. 1. 3 , page 494] l Density-based – A cluster is a dense region of points, which is separated by low -density regions, from other regions of high density. – Used when the clusters are irregular or intertwined, and when noise and outliers are present. 6 density-based clusters 2010/5/7 16
![Types of Clusters Conceptual Clusters Chapter 8 1 3 page 495 l Shared Types of Clusters: Conceptual Clusters [Chapter 8. 1. 3 , page 495] l Shared](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-16.jpg)
Types of Clusters: Conceptual Clusters [Chapter 8. 1. 3 , page 495] l Shared Property or Conceptual Clusters – Finds clusters that share some common property or represent a particular concept. . 2 Overlapping Circles 2010/5/7 17
![Types of Clusters Objective Function optional l Clusters Defined by an Objective Function Types of Clusters: Objective Function [optional] l Clusters Defined by an Objective Function –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-17.jpg)
Types of Clusters: Objective Function [optional] l Clusters Defined by an Objective Function – Finds clusters that minimize or maximize an objective function. – Enumerate all possible ways of dividing the points into clusters and evaluate the `goodness' of each potential set of clusters by using the given objective function. (NP Hard) – Can have global or local objectives. u Hierarchical clustering algorithms typically have local objectives u Partitional algorithms typically have global objectives – A variation of the global objective function approach is to fit the data to a parameterized model. u Parameters for the model are determined from the data. Mixture models assume that the data is a ‘mixture' of a number of statistical distributions. u 2010/5/7 18
![Types of Clusters Objective Function optional l Map the clustering problem to a Types of Clusters: Objective Function … [optional] l Map the clustering problem to a](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-18.jpg)
Types of Clusters: Objective Function … [optional] l Map the clustering problem to a different domain and solve a related problem in that domain – Proximity matrix defines a weighted graph, where the nodes are the points being clustered, and the weighted edges represent the proximities between points – Clustering is equivalent to breaking the graph into connected components, one for each cluster. – Want to minimize the edge weight between clusters and maximize the edge weight within clusters 2010/5/7 19
![Characteristics of the Input Data Are Important optional l l Size Scale Normalization Characteristics of the Input Data Are Important [optional] l l Size Scale – Normalization](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-19.jpg)
Characteristics of the Input Data Are Important [optional] l l Size Scale – Normalization l Sparseness – Dictates type of similarity (proximity- type or density- type of measure) l Attribute type – Dictates type of similarity l Type of Data – Dictates type of similarity l l l Dimensionality Noise and Outliers Type of Distribution 2010/5/7 20

Clustering Algorithms l K-means and its variants l Hierarchical clustering l Density-based clustering 2010/5/7 [Chapter 8. 2 , page 496] 21
![Kmeans Clustering Chapter 8 2 1 page 497 l Partitional clustering approach l K-means Clustering [Chapter 8. 2. 1 , page 497] l Partitional clustering approach l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-21.jpg)
K-means Clustering [Chapter 8. 2. 1 , page 497] l Partitional clustering approach l Each cluster is associated with a centroid (center point) l Each point is assigned to the cluster with the closest centroid l Number of clusters, K, must be specified l The basic algorithm is very simple 2010/5/7 22
![Kmeans Clustering Details l Chapter 8 2 1 Initial centroids are often chosen K-means Clustering – Details l [Chapter 8. 2. 1] Initial centroids are often chosen](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-22.jpg)
K-means Clustering – Details l [Chapter 8. 2. 1] Initial centroids are often chosen randomly. – Clusters produced vary from one run to another. l The centroid is (typically) the mean of the points in the cluster. l ‘Closeness’ is measured by Euclidean distance, cosine similarity, correlation, etc. l K-means will converge for common similarity measures mentioned above. l Most of the convergence happens in the first few iterations. – l Complexity is O( n * K * I * d ) – 2010/5/7 Often the stopping condition is changed to ‘Until relatively few points change clusters’ n = number of points, K = number of clusters, I = number of iterations, d = number of attributes 23
![Two different Kmeans Clusterings Chapter 8 2 1 Original Points Optimal Clustering 201057 Suboptimal Two different K-means Clusterings [Chapter 8. 2. 1] Original Points Optimal Clustering 2010/5/7 Sub-optimal](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-23.jpg)
Two different K-means Clusterings [Chapter 8. 2. 1] Original Points Optimal Clustering 2010/5/7 Sub-optimal Clustering 24
![Importance of Choosing Initial Centroids optional 201057 25 Importance of Choosing Initial Centroids [optional] 2010/5/7 25](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-24.jpg)
Importance of Choosing Initial Centroids [optional] 2010/5/7 25
![Importance of Choosing Initial Centroids optional 201057 26 Importance of Choosing Initial Centroids [optional] 2010/5/7 26](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-25.jpg)
Importance of Choosing Initial Centroids [optional] 2010/5/7 26

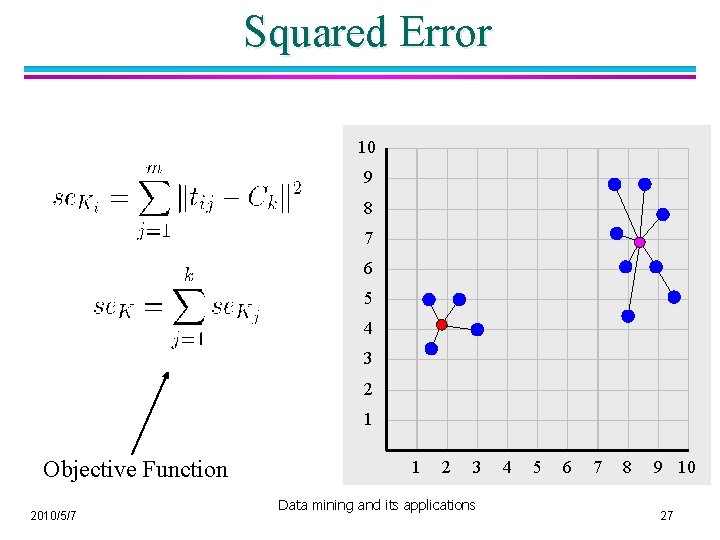
Squared Error 10 9 8 7 6 5 4 3 2 1 Objective Function 2010/5/7 1 2 3 Data mining and its applications 4 5 6 7 8 9 10 27
![Evaluating Kmeans Clusters Chapter 8 2 1 page 499 l Most common measure Evaluating K-means Clusters [Chapter 8. 2. 1 , page 499] l Most common measure](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-27.jpg)
Evaluating K-means Clusters [Chapter 8. 2. 1 , page 499] l Most common measure is Sum of Squared Error (SSE) – For each point, the error is the distance to the nearest cluster – To get SSE, we square these errors and sum them. – x is a data point in cluster Ci and mi is the representative point for cluster Ci u l l can show that mi corresponds to the center (mean) of the cluster Given two clusterings, we can choose the one with the smallest error One easy way to reduce SSE is to increase K, the number of clusters – 2010/5/7 A good clustering with smaller K can have a lower SSE than a poor clustering with higher K 28
![Importance of Choosing Initial Centroids Chapter 8 2 1 page 501 201057 Importance of Choosing Initial Centroids … [Chapter 8. 2. 1 , page 501] 2010/5/7](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-28.jpg)
Importance of Choosing Initial Centroids … [Chapter 8. 2. 1 , page 501] 2010/5/7 29
![Importance of Choosing Initial Centroids Chapter 8 2 1 page 501 201057 Importance of Choosing Initial Centroids … [Chapter 8. 2. 1 , page 501] 2010/5/7](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-29.jpg)
Importance of Choosing Initial Centroids … [Chapter 8. 2. 1 , page 501] 2010/5/7 30

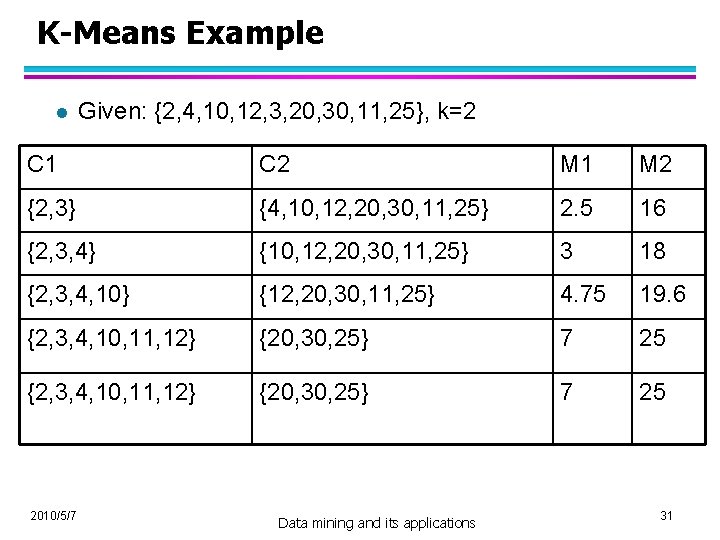
K-Means Example l Given: {2, 4, 10, 12, 3, 20, 30, 11, 25}, k=2 C 1 C 2 M 1 M 2 {2, 3} {4, 10, 12, 20, 30, 11, 25} 2. 5 16 {2, 3, 4} {10, 12, 20, 30, 11, 25} 3 18 {2, 3, 4, 10} {12, 20, 30, 11, 25} 4. 75 19. 6 {2, 3, 4, 10, 11, 12} {20, 30, 25} 7 25 2010/5/7 Data mining and its applications 31
![Problems with Selecting Initial Points Chapter 8 2 1 page 503 l 201057 Problems with Selecting Initial Points [Chapter 8. 2. 1 , page 503] l 2010/5/7](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-31.jpg)
Problems with Selecting Initial Points [Chapter 8. 2. 1 , page 503] l 2010/5/7 If there are K ‘real’ clusters then the chance of selecting one centroid from each cluster is small. – – Chance is relatively small when K is large – – For example, if K = 10, then probability = 10!/1010 = 0. 00036 – Consider an example of five pairs of clusters If clusters are the same size, n, then Sometimes the initial centroids will readjust themselves in ‘right’ way, and sometimes they don’t 32
![10 Clusters Example optional Starting with two initial centroids in one cluster of each 10 Clusters Example [optional] Starting with two initial centroids in one cluster of each](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-32.jpg)
10 Clusters Example [optional] Starting with two initial centroids in one cluster of each pair of clusters 2010/5/7 33
![10 Clusters Example Optional Starting with two initial centroids in one cluster of each 10 Clusters Example [Optional] Starting with two initial centroids in one cluster of each](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-33.jpg)
10 Clusters Example [Optional] Starting with two initial centroids in one cluster of each pair of clusters 2010/5/7 34
![10 Clusters Example optional Starting with some pairs of clusters having three initial centroids 10 Clusters Example [optional] Starting with some pairs of clusters having three initial centroids,](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-34.jpg)
10 Clusters Example [optional] Starting with some pairs of clusters having three initial centroids, while other have only one. 2010/5/7 35
![10 Clusters Example optional Starting with some pairs of clusters having three initial centroids 10 Clusters Example [optional] Starting with some pairs of clusters having three initial centroids,](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-35.jpg)
10 Clusters Example [optional] Starting with some pairs of clusters having three initial centroids, while other have only one. 2010/5/7 36
![Solutions to Initial Centroids Problem Chapter 8 2 1 page 504 l Multiple Solutions to Initial Centroids Problem [Chapter 8. 2. 1 , page 504] l Multiple](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-36.jpg)
Solutions to Initial Centroids Problem [Chapter 8. 2. 1 , page 504] l Multiple runs – Helps, but probability is not on your side Sample and use hierarchical clustering to determine initial centroids l Select more than k initial centroids and then select among these initial centroids l – Select most widely separated Postprocessing l Bisecting K-means l – Not as susceptible to initialization issues 2010/5/7 37
![Handling Empty Clusters Chapter 8 2 2 page 506 l Basic Kmeans algorithm Handling Empty Clusters [Chapter 8. 2. 2 , page 506] l Basic K-means algorithm](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-37.jpg)
Handling Empty Clusters [Chapter 8. 2. 2 , page 506] l Basic K-means algorithm can yield empty clusters l Several strategies – Choose the point that contributes most to SSE – Choose a point from the cluster with the highest SSE – If there are several empty clusters, the above can be repeated several times. 2010/5/7 38
![Updating Centers Incrementally Chapter 8 2 2 page 508 l In the basic Updating Centers Incrementally [Chapter 8. 2. 2 , page 508] l In the basic](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-38.jpg)
Updating Centers Incrementally [Chapter 8. 2. 2 , page 508] l In the basic K-means algorithm, centroids are updated after all points are assigned to a centroid l An alternative is to update the centroids after each assignment (incremental approach) – – – 2010/5/7 Never get an empty cluster Can use “weights” to change the impact Each assignment updates zero or two centroids More expensive Introduces an order dependency 39
![Preprocessing and Postprocessing Chapter 8 2 2 l Preprocessing Normalize the data Pre-processing and Post-processing [Chapter 8. 2. 2] l Pre-processing – Normalize the data –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-39.jpg)
Pre-processing and Post-processing [Chapter 8. 2. 2] l Pre-processing – Normalize the data – Eliminate outliers l Post-processing – To decrease the total SSE u. Split ‘loose’ clusters, i. e. , clusters with relatively high SSE u. Introduce a ne cluster centroid – To decrease the number of clusters u. Merge clusters that are ‘close’ or result in the smallest increase in total SSE u. Eliminate small clusters that may represent outliers – Can use these steps during the clustering process 2010/5/7 40
![Bisecting Kmeans l Bisecting Kmeans algorithm 201057 Chapter 8 2 3 page 509 Bisecting K-means l Bisecting K-means algorithm – 2010/5/7 [Chapter 8. 2. 3, page 509]](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-40.jpg)
Bisecting K-means l Bisecting K-means algorithm – 2010/5/7 [Chapter 8. 2. 3, page 509] Variant of K-means that can produce a partitional or a hierarchical clustering 41
![Bisecting Kmeans Example 201057 Chapter 8 2 3 42 Bisecting K-means Example 2010/5/7 [Chapter 8. 2. 3] 42](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-41.jpg)
Bisecting K-means Example 2010/5/7 [Chapter 8. 2. 3] 42
![Limitations of Kmeans l Chapter 8 2 4 Kmeans has problems when clusters are Limitations of K-means l [Chapter 8. 2. 4] K-means has problems when clusters are](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-42.jpg)
Limitations of K-means l [Chapter 8. 2. 4] K-means has problems when clusters are of differing – Sizes – Densities – Non-globular shapes l K-means has problems when the data contains outliers. 2010/5/7 43
![Limitations of Kmeans Differing Sizes Chapter 8 2 4 page 511 Original Points Limitations of K-means: Differing Sizes [Chapter 8. 2. 4 , page 511] Original Points](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-43.jpg)
Limitations of K-means: Differing Sizes [Chapter 8. 2. 4 , page 511] Original Points 2010/5/7 K-means (3 Clusters) 44
![Limitations of Kmeans Differing Density Chapter 8 2 4 page 511 Original Points Limitations of K-means: Differing Density [Chapter 8. 2. 4 , page 511] Original Points](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-44.jpg)
Limitations of K-means: Differing Density [Chapter 8. 2. 4 , page 511] Original Points 2010/5/7 K-means (3 Clusters) 45
![Limitations of Kmeans Nonglobular Shapes Chapter 8 2 4 page 511 Original Points Limitations of K-means: Non-globular Shapes [Chapter 8. 2. 4 , page 511] Original Points](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-45.jpg)
Limitations of K-means: Non-globular Shapes [Chapter 8. 2. 4 , page 511] Original Points 2010/5/7 K-means (2 Clusters) 46
![Overcoming Kmeans Limitations Chapter 8 2 4optional Original Points Kmeans Clusters One solution is Overcoming K-means Limitations [Chapter 8. 2. 4_optional] Original Points K-means Clusters One solution is](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-46.jpg)
Overcoming K-means Limitations [Chapter 8. 2. 4_optional] Original Points K-means Clusters One solution is to use many clusters. Find parts of clusters, but need to put together. 2010/5/7 47
![Overcoming Kmeans LimitationsChapter 8 2 4optional Original Points 201057 Kmeans Clusters 48 Overcoming K-means Limitations[Chapter 8. 2. 4_optional] Original Points 2010/5/7 K-means Clusters 48](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-47.jpg)
Overcoming K-means Limitations[Chapter 8. 2. 4_optional] Original Points 2010/5/7 K-means Clusters 48
![Overcoming Kmeans Limitations Chapter 8 2 4optional Original Points 201057 Kmeans Clusters 49 Overcoming K-means Limitations [Chapter 8. 2. 4_optional] Original Points 2010/5/7 K-means Clusters 49](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-48.jpg)
Overcoming K-means Limitations [Chapter 8. 2. 4_optional] Original Points 2010/5/7 K-means Clusters 49
![Hierarchical Clustering Chapter 8 3 page 515 516 Produces a set of nested Hierarchical Clustering [Chapter 8. 3 , page 515 -516] Produces a set of nested](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-49.jpg)
Hierarchical Clustering [Chapter 8. 3 , page 515 -516] Produces a set of nested clusters organized as a hierarchical tree l Can be visualized as a dendrogram l – A tree like diagram that records the sequences of merges (or splits) dendrogram 2010/5/7 nested cluster diagram 50
![Strengths of Hierarchical Clustering chapter 8 3 l Do not have to assume any Strengths of Hierarchical Clustering [chapter 8. 3] l Do not have to assume any](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-50.jpg)
Strengths of Hierarchical Clustering [chapter 8. 3] l Do not have to assume any particular number of clusters – Any desired number of clusters can be obtained by ‘cutting’ the dendogram at the proper level l They may correspond to meaningful taxonomies – Example in biological sciences (e. g. , animal kingdom, phylogeny reconstruction, …) 2010/5/7 51
![Hierarchical Clustering l Chapter 8 3 page 515 Two main types of hierarchical Hierarchical Clustering l [Chapter 8. 3 , page 515] Two main types of hierarchical](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-51.jpg)
Hierarchical Clustering l [Chapter 8. 3 , page 515] Two main types of hierarchical clustering – Agglomerative: u Start with the points as individual clusters At each step, merge the closest pair of clusters until only one cluster (or k clusters) left u – Divisive: u Start with one, all-inclusive cluster At each step, split a cluster until each cluster contains a point (or there are k clusters) u l Traditional hierarchical algorithms use a similarity or distance matrix – Merge or split one cluster at a time 2010/5/7 52
![Agglomerative Clustering Algorithm Chapter 8 3 1 page 516 l More popular hierarchical Agglomerative Clustering Algorithm [Chapter 8. 3. 1 , page 516] l More popular hierarchical](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-52.jpg)
Agglomerative Clustering Algorithm [Chapter 8. 3. 1 , page 516] l More popular hierarchical clustering technique l Basic algorithm is straightforward 1. 2. 3. 4. 5. 6. l Let each data point be a cluster Repeat Merge the two closest clusters Update the proximity matrix Until only a single cluster remains Key operation is the computation of the proximity of two clusters – 2010/5/7 Compute the proximity matrix Different approaches to defining the distance between clusters distinguish the different algorithms 53
![Starting Situation l optional Start with clusters of individual points and a proximity matrix Starting Situation l [optional] Start with clusters of individual points and a proximity matrix](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-53.jpg)
Starting Situation l [optional] Start with clusters of individual points and a proximity matrix p 1 p 2 p 3 p 4 p 5 . . . p 1 p 2 p 3 p 4 p 5. . . 2010/5/7 Proximity Matrix 54
![Intermediate Situation l optional After some merging steps we have some clusters C 1 Intermediate Situation l [optional] After some merging steps, we have some clusters C 1](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-54.jpg)
Intermediate Situation l [optional] After some merging steps, we have some clusters C 1 C 2 C 3 C 4 C 5 Proximity Matrix C 1 C 2 2010/5/7 C 5 55
![Intermediate Situation l optional We want to merge the two closest clusters C 2 Intermediate Situation l [optional] We want to merge the two closest clusters (C 2](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-55.jpg)
Intermediate Situation l [optional] We want to merge the two closest clusters (C 2 and C 5) and update the proximity matrix. C 1 C 2 C 3 C 4 C 5 Proximity Matrix C 1 C 2 2010/5/7 C 5 56
![After Merging l optional The question is How do we update the proximity matrix After Merging l [optional] The question is “How do we update the proximity matrix?](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-56.jpg)
After Merging l [optional] The question is “How do we update the proximity matrix? ” C 1 C 2 U C 5 C 3 C 4 ? ? ? C 3 ? C 4 ? Proximity Matrix C 1 C 2 U C 5 2010/5/7 57
![How to Define InterCluster Similarity Chapter 8 3 1 page 517 p 1 How to Define Inter-Cluster Similarity [Chapter 8. 3. 1 , page 517] p 1](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-57.jpg)
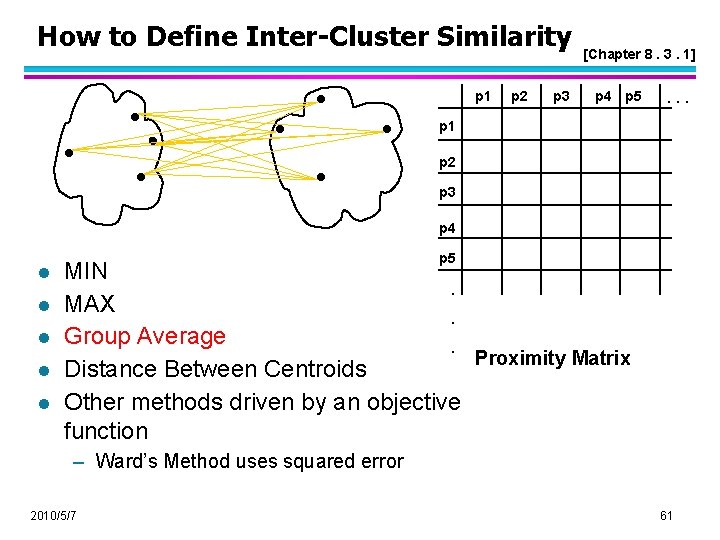
How to Define Inter-Cluster Similarity [Chapter 8. 3. 1 , page 517] p 1 Similarity? p 2 p 3 p 4 p 5 . . . p 1 p 2 p 3 p 4 l l l p 5 MIN. MAX. Group Average. Proximity Matrix Distance Between Centroids Other methods driven by an objective function – Ward’s Method uses squared error 2010/5/7 58
![How to Define InterCluster SimilarityChapter 8 3 1 p 1 p 2 p 3 How to Define Inter-Cluster Similarity[Chapter 8. 3. 1] p 1 p 2 p 3](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-58.jpg)
How to Define Inter-Cluster Similarity[Chapter 8. 3. 1] p 1 p 2 p 3 p 4 p 5 . . . p 1 p 2 p 3 p 4 l l l p 5 MIN. MAX. Group Average. Proximity Matrix Distance Between Centroids Other methods driven by an objective function – Ward’s Method uses squared error 2010/5/7 59
![How to Define InterCluster SimilarityChapter 8 3 1 p 1 p 2 p 3 How to Define Inter-Cluster Similarity[Chapter 8. 3. 1] p 1 p 2 p 3](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-59.jpg)
How to Define Inter-Cluster Similarity[Chapter 8. 3. 1] p 1 p 2 p 3 p 4 p 5 . . . p 1 p 2 p 3 p 4 l l l p 5 MIN. MAX. Group Average. Proximity Matrix Distance Between Centroids Other methods driven by an objective function – Ward’s Method uses squared error 2010/5/7 60

How to Define Inter-Cluster Similarity p 1 p 2 p 3 [Chapter 8. 3. 1] p 4 p 5 . . . p 1 p 2 p 3 p 4 l l l p 5 MIN. MAX. Group Average. Proximity Matrix Distance Between Centroids Other methods driven by an objective function – Ward’s Method uses squared error 2010/5/7 61
![How to Define InterCluster Similarity Chapter 8 3 1 p 1 p 2 p How to Define Inter-Cluster Similarity [Chapter 8. 3. 1] p 1 p 2 p](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-61.jpg)
How to Define Inter-Cluster Similarity [Chapter 8. 3. 1] p 1 p 2 p 3 p 4 p 5 . . . p 1 p 2 p 3 p 4 l l l p 5 MIN. MAX. Group Average. Proximity Matrix Distance Between Centroids Other methods driven by an objective function – Ward’s Method uses squared error 2010/5/7 62
![Cluster Similarity MIN or Single Link Chapter 8 3 2 page 519 l Cluster Similarity: MIN or Single Link [Chapter 8. 3. 2 , page 519] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-62.jpg)
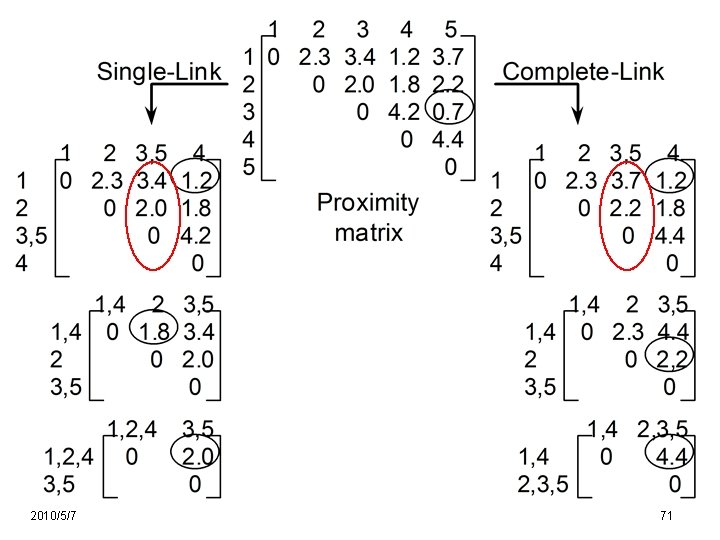
Cluster Similarity: MIN or Single Link [Chapter 8. 3. 2 , page 519] l Similarity of two clusters is based on the two most similar (closest) points in the different clusters – Determined by one pair of points, i. e. , by one link in the proximity graph. Ex. Euclidean Distance 2010/5/7 1 2 3 4 5 63
![Hierarchical Clustering MIN Chapter 8 3 2 page 520 1 3 5 2 Hierarchical Clustering: MIN [Chapter 8. 3. 2 , page 520] 1 3 5 2](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-63.jpg)
Hierarchical Clustering: MIN [Chapter 8. 3. 2 , page 520] 1 3 5 2 1 2 3 4 6 4 Nested Clusters 2010/5/7 5 Dendrogram 64
![Strength of MIN Original Points Chapter 8 3 2 Two Clusters Can Strength of MIN Original Points [Chapter 8. 3. 2 ] Two Clusters • Can](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-64.jpg)
Strength of MIN Original Points [Chapter 8. 3. 2 ] Two Clusters • Can handle non-elliptical shapes 2010/5/7 65
![Limitations of MIN Original Points Chapter 8 3 2 Two Clusters Sensitive to Limitations of MIN Original Points [Chapter 8. 3. 2] Two Clusters • Sensitive to](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-65.jpg)
Limitations of MIN Original Points [Chapter 8. 3. 2] Two Clusters • Sensitive to noise and outliers 2010/5/7 66
![Cluster Similarity MAX or Complete Linkage Chapter 8 3 2 page 520 l Cluster Similarity: MAX or Complete Linkage [Chapter 8. 3. 2 , page 520] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-66.jpg)
Cluster Similarity: MAX or Complete Linkage [Chapter 8. 3. 2 , page 520] l Similarity of two clusters is based on the two least similar (most distant) points in the different clusters – Determined by all pairs of points in the two clusters 1 2010/5/7 2 3 4 5 67
![Hierarchical Clustering MAX Chapter 8 3 2 page 521 4 1 2 5 Hierarchical Clustering: MAX [Chapter 8. 3. 2 , page 521] 4 1 2 5](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-67.jpg)
Hierarchical Clustering: MAX [Chapter 8. 3. 2 , page 521] 4 1 2 5 5 2 3 3 6 1 4 Nested Clusters 2010/5/7 Dendrogram 68
![Strength of MAX Original Points Chapter 8 3 2 Two Clusters Less susceptible Strength of MAX Original Points [Chapter 8. 3. 2] Two Clusters • Less susceptible](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-68.jpg)
Strength of MAX Original Points [Chapter 8. 3. 2] Two Clusters • Less susceptible to noise and outliers 2010/5/7 69
![Limitations of MAX Original Points Chapter 8 3 2 Two Clusters Tends Limitations of MAX Original Points [Chapter 8. 3. 2 ] Two Clusters • Tends](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-69.jpg)
Limitations of MAX Original Points [Chapter 8. 3. 2 ] Two Clusters • Tends to break large clusters • Biased towards globular clusters 2010/5/7 70

2010/5/7 71
![Cluster Similarity Group Average Chapter 8 3 2 page 521 l Proximity of Cluster Similarity: Group Average [Chapter 8. 3. 2 , page 521] l Proximity of](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-71.jpg)
Cluster Similarity: Group Average [Chapter 8. 3. 2 , page 521] l Proximity of two clusters is the average of pairwise proximity between points in the two clusters. l Need to use average connectivity for scalability since total proximity favors large clusters 1 2010/5/7 2 3 4 5 72
![Hierarchical Clustering Group Average Chapter 8 3 2 page 523 5 4 1 Hierarchical Clustering: Group Average [Chapter 8. 3. 2 , page 523] 5 4 1](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-72.jpg)
Hierarchical Clustering: Group Average [Chapter 8. 3. 2 , page 523] 5 4 1 2 5 2 3 6 1 4 3 Nested Clusters 2010/5/7 Dendrogram 73
![Hierarchical Clustering Group Average Chapter 8 3 2 l Compromise between Single and Complete Hierarchical Clustering: Group Average [Chapter 8. 3. 2] l Compromise between Single and Complete](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-73.jpg)
Hierarchical Clustering: Group Average [Chapter 8. 3. 2] l Compromise between Single and Complete Link l Strengths – Less susceptible to noise and outliers Limitations l – Biased towards globular clusters 2010/5/7 74
![Cluster Similarity Wards Method Chapter 8 3 2 page 523 l Similarity of Cluster Similarity: Ward’s Method [Chapter 8. 3. 2 , page 523] l Similarity of](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-74.jpg)
Cluster Similarity: Ward’s Method [Chapter 8. 3. 2 , page 523] l Similarity of two clusters is based on the increase in squared error when two clusters are merged – Similar to group average if distance between points is distance squared l Less susceptible to noise and outliers l Biased towards globular clusters l Hierarchical analogue of K-means – Can be used to initialize K-means 2010/5/7 75

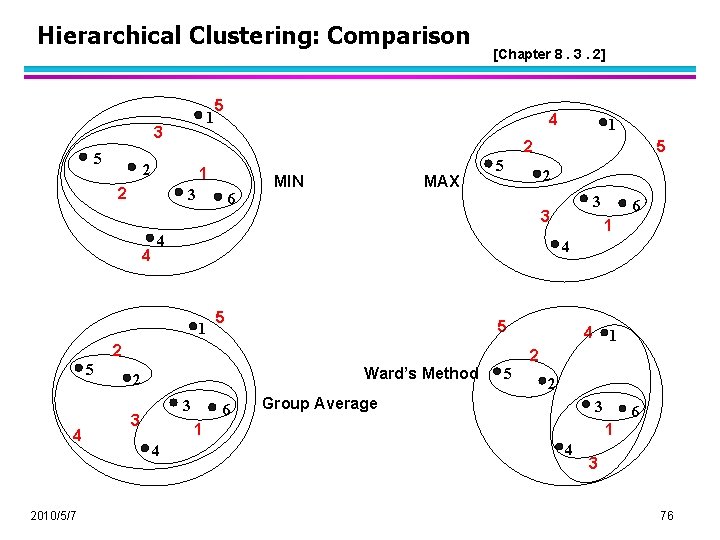
Hierarchical Clustering: Comparison 1 3 5 5 1 3 4 4 6 MIN MAX 5 2010/5/7 2 5 1 5 Ward’s Method 3 6 4 1 2 5 2 Group Average 3 1 4 6 4 2 3 3 3 2 4 5 4 1 5 1 2 2 2 [Chapter 8. 3. 2] 6 1 4 3 76
![Hierarchical Clustering Time and Space requirements Chapter 8 3 4 l ON 2 space Hierarchical Clustering: Time and Space requirements [Chapter 8. 3. 4] l O(N 2) space](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-76.jpg)
Hierarchical Clustering: Time and Space requirements [Chapter 8. 3. 4] l O(N 2) space since it uses the proximity matrix. – N is the number of points. l O(N 3) time in many cases – There are N steps and at each step the size, N 2, proximity matrix must be updated and searched – Complexity can be reduced to O(N 2 log(N) ) time for some approaches 2010/5/7 77
![Hierarchical Clustering Problems and Limitations Chapter 8 3 5 l Once a decision is Hierarchical Clustering: Problems and Limitations [Chapter 8. 3. 5] l Once a decision is](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-77.jpg)
Hierarchical Clustering: Problems and Limitations [Chapter 8. 3. 5] l Once a decision is made to combine two clusters, it cannot be undone l No objective function is directly minimized l Different schemes have problems with one or more of the following: – Sensitivity to noise and outliers – Difficulty handling different sized clusters and convex shapes – Breaking large clusters 2010/5/7 78

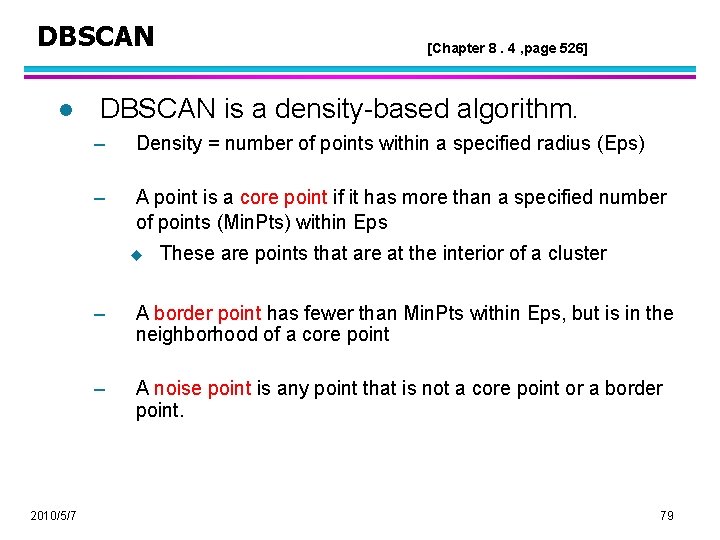
DBSCAN l DBSCAN is a density-based algorithm. – Density = number of points within a specified radius (Eps) – A point is a core point if it has more than a specified number of points (Min. Pts) within Eps u 2010/5/7 [Chapter 8. 4 , page 526] These are points that are at the interior of a cluster – A border point has fewer than Min. Pts within Eps, but is in the neighborhood of a core point – A noise point is any point that is not a core point or a border point. 79
![DBSCAN Core Border and Noise Points Chapter 8 4 1 page 528 201057 DBSCAN: Core, Border, and Noise Points [Chapter 8. 4. 1 , page 528] 2010/5/7](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-79.jpg)
DBSCAN: Core, Border, and Noise Points [Chapter 8. 4. 1 , page 528] 2010/5/7 80
![DBSCAN Algorithm Chapter 8 4 2 page 528 Eliminate noise points l Perform DBSCAN Algorithm [Chapter 8. 4. 2 , page 528] Eliminate noise points l Perform](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-80.jpg)
DBSCAN Algorithm [Chapter 8. 4. 2 , page 528] Eliminate noise points l Perform clustering on the remaining points l 2010/5/7 81
![DBSCAN Core Border and Noise Points Chapter 8 4 2 page 530 Original DBSCAN: Core, Border and Noise Points [Chapter 8. 4. 2 , page 530] Original](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-81.jpg)
DBSCAN: Core, Border and Noise Points [Chapter 8. 4. 2 , page 530] Original Points Point types: core, border and noise Eps = 10, Min. Pts = 4 2010/5/7 82
![When DBSCAN Works Well Original Points optional Clusters Resistant to Noise Can When DBSCAN Works Well Original Points [optional] Clusters • Resistant to Noise • Can](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-82.jpg)
When DBSCAN Works Well Original Points [optional] Clusters • Resistant to Noise • Can handle clusters of different shapes and sizes 2010/5/7 83
![When DBSCAN Does NOT Work Well optional Min Pts4 Eps9 75 Original Points When DBSCAN Does NOT Work Well [optional] (Min. Pts=4, Eps=9. 75). Original Points •](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-83.jpg)
When DBSCAN Does NOT Work Well [optional] (Min. Pts=4, Eps=9. 75). Original Points • Varying densities • High-dimensional data 2010/5/7 (Min. Pts=4, Eps=9. 92) 84
![DBSCAN Determining EPS and Min Pts l l l 201057 optional Idea is that DBSCAN: Determining EPS and Min. Pts l l l 2010/5/7 [optional] Idea is that](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-84.jpg)
DBSCAN: Determining EPS and Min. Pts l l l 2010/5/7 [optional] Idea is that for points in a cluster, their kth nearest neighbors are at roughly the same distance Noise points have the kth nearest neighbor at farther distance So, plot sorted distance of every point to its kth nearest neighbor 85
![Different Aspects of Cluster Validation Chapter 8 5 1 page 533 1 Determining Different Aspects of Cluster Validation [Chapter 8. 5. 1 , page 533] 1. Determining](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-85.jpg)
Different Aspects of Cluster Validation [Chapter 8. 5. 1 , page 533] 1. Determining the clustering tendency of a set of data, i. e. , distinguishing whether non-random structure actually exists in the data. 2. Comparing the results of a cluster analysis to externally known results, e. g. , to externally given class labels. 3. Evaluating how well the results of a cluster analysis fit the data without reference to external information. - Use only the data 4. Comparing the results of two different sets of cluster analyses to determine which is better. 5. Determining the ‘correct’ number of clusters. For 2, 3, and 4, we can further distinguish whether we want to evaluate the entire clustering or just individual clusters. 2010/5/7 86
![Internal Measures Cohesion and Separation Chapter 8 5 2 page 536 l Cluster Internal Measures: Cohesion and Separation [Chapter 8. 5. 2 , page 536] l Cluster](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-86.jpg)
Internal Measures: Cohesion and Separation [Chapter 8. 5. 2 , page 536] l Cluster Cohesion: Measures how closely related are objects in a cluster – Example: SSE l Cluster Separation: Measure how distinct or wellseparated a cluster is from other clusters l Example: Squared Error – Cohesion is measured by the within cluster sum of squares (SSE) – Separation is measured by the between cluster sum of squares – Where |Ci| is the size of cluster i 2010/5/7 87
![Internal Measures Cohesion and Separation Chapter 8 5 2 l Example SSE BSS Internal Measures: Cohesion and Separation [Chapter 8. 5. 2] l Example: SSE – BSS](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-87.jpg)
Internal Measures: Cohesion and Separation [Chapter 8. 5. 2] l Example: SSE – BSS + WSS = constant 1 m 2 3 4 m 2 5 K=1 cluster: K=2 clusters: 2010/5/7 88
![Internal Measures Cohesion and Separation Chapter 8 5 2 l A proximity graph based Internal Measures: Cohesion and Separation [Chapter 8. 5. 2] l A proximity graph based](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-88.jpg)
Internal Measures: Cohesion and Separation [Chapter 8. 5. 2] l A proximity graph based approach can also be used for cohesion and separation. – Cluster cohesion is the sum of the weight of all links within a cluster. – Cluster separation is the sum of the weights between nodes in the cluster and nodes outside the cluster. cohesion 2010/5/7 separation 89
![Internal Measures Silhouette Coefficient Chapter 8 5 2 page 541 l l Silhouette Internal Measures: Silhouette Coefficient [Chapter 8. 5. 2 , page 541] l l Silhouette](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-89.jpg)
Internal Measures: Silhouette Coefficient [Chapter 8. 5. 2 , page 541] l l Silhouette Coefficient combine ideas of both cohesion and separation, but for individual points, as well as clusters and clusterings For an individual point, i – Calculate a = average distance of i to the points in its cluster – Calculate b = min (average distance of i to points in another cluster) – The silhouette coefficient for a point is then given by s = 1 – a/b if a < b, (or s = b/a - 1 if a b, not the usual case) – Typically between 0 and 1. – The closer to 1 the better. l Can calculate the Average Silhouette width for a clustering 2010/5/7 90
![Using Similarity Matrix for Cluster Validation Chapter 8 5 3 page 544 l Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 544] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-90.jpg)
Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 544] l Order the similarity matrix with respect to cluster labels and inspect visually. 2010/5/7 91
![Using Similarity Matrix for Cluster Validation Chapter 8 5 3 page 545 l Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-91.jpg)
Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l Clusters in random data are not so crisp DBSCAN 2010/5/7 92
![Using Similarity Matrix for Cluster Validation Chapter 8 5 3 page 545 l Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-92.jpg)
Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l Clusters in random data are not so crisp K-means 2010/5/7 93
![Using Similarity Matrix for Cluster Validation Chapter 8 5 3 page 545 l Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-93.jpg)
Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3 , page 545] l Clusters in random data are not so crisp Complete Link 2010/5/7 94
![Using Similarity Matrix for Cluster Validation Chapter 8 5 3 DBSCAN 201057 95 Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3] DBSCAN 2010/5/7 95](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-94.jpg)
Using Similarity Matrix for Cluster Validation [Chapter 8. 5. 3] DBSCAN 2010/5/7 95
![Internal Measures SSE optional l Clusters in more complicated figures arent well separated l Internal Measures: SSE [optional] l Clusters in more complicated figures aren’t well separated l](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-95.jpg)
Internal Measures: SSE [optional] l Clusters in more complicated figures aren’t well separated l Internal Index: Used to measure the goodness of a clustering structure without respect to external information – SSE l l SSE is good for comparing two clusterings or two clusters (average SSE). Can also be used to estimate the number of clusters 2010/5/7 96
![Internal Measures SSE l optional SSE curve for a more complicated data set SSE Internal Measures: SSE l [optional] SSE curve for a more complicated data set SSE](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-96.jpg)
Internal Measures: SSE l [optional] SSE curve for a more complicated data set SSE of clusters found using K-means 2010/5/7 97

Framework for Cluster Validity l Need a framework to interpret any measure. – l For example, if our measure of evaluation has the value, 10, is that good, fair, or poor? Statistics provide a framework for cluster validity – The more “atypical” a clustering result is, the more likely it represents valid structure in the data – Can compare the values of an index that result from random data or clusterings to those of a clustering result. u – l If the value of the index is unlikely, then the cluster results are valid These approaches are more complicated and harder to understand. For comparing the results of two different sets of cluster analyses, a framework is less necessary. – 2010/5/7 [Chapter 8. 5. 3] However, there is the question of whether the difference between two index values is significant 98
![Cluster Validity l Chapter 8 5 7 page 548 For supervised classification we Cluster Validity l [Chapter 8. 5. 7 , page 548] For supervised classification we](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-98.jpg)
Cluster Validity l [Chapter 8. 5. 7 , page 548] For supervised classification we have a variety of measures to evaluate how good our model is – Accuracy, precision, recall l For cluster analysis, the analogous question is how to evaluate the “goodness” of the resulting clusters? l But “clusters are in the eye of the beholder”! l Then why do we want to evaluate them? – – 2010/5/7 To avoid finding patterns in noise To compare clustering algorithms To compare two sets of clusters To compare two clusters 99
![Clusters found in Random Data Random Points Kmeans 201057 optional DBSCAN Complete Link 100 Clusters found in Random Data Random Points K-means 2010/5/7 [optional] DBSCAN Complete Link 100](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-99.jpg)
Clusters found in Random Data Random Points K-means 2010/5/7 [optional] DBSCAN Complete Link 100

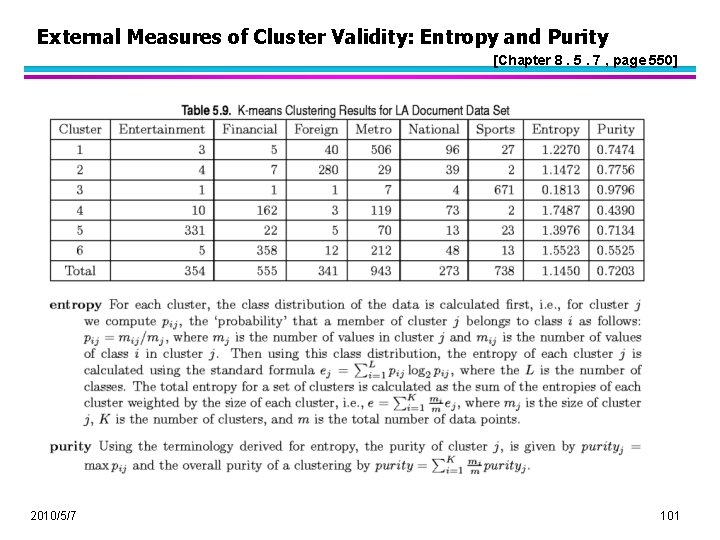
External Measures of Cluster Validity: Entropy and Purity [Chapter 8. 5. 7 , page 550] 2010/5/7 101
![Measures of Cluster Validity l Chapter 8 5 7 Numerical measures that are applied Measures of Cluster Validity l [Chapter 8. 5. 7] Numerical measures that are applied](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-101.jpg)
Measures of Cluster Validity l [Chapter 8. 5. 7] Numerical measures that are applied to judge various aspects of cluster validity, are classified into the following three types. – External Index: Used to measure the extent to which cluster labels match externally supplied class labels. u Entropy – Internal Index: Used to measure the goodness of a clustering structure without respect to external information. u Sum of Squared Error (SSE) – Relative Index: Used to compare two different clusterings or clusters. u l Often an external or internal index is used for this function, e. g. , SSE or entropy Sometimes these are referred to as criteria instead of indices – However, sometimes criterion is the general strategy and index is the numerical measure that implements the criterion. 2010/5/7 102
![Measuring Cluster Validity Via Correlation Chapter 8 5 7 l l Two matrices Measuring Cluster Validity Via Correlation [Chapter 8. 5. 7] l l Two matrices –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-102.jpg)
Measuring Cluster Validity Via Correlation [Chapter 8. 5. 7] l l Two matrices – Proximity Matrix – “Incidence” Matrix l 2010/5/7 One row and one column for each data point u An entry is 1 if the associated pair of points belong to the same cluster u An entry is 0 if the associated pair of points belongs to different clusters Compute the correlation between the two matrices – l u Since the matrices are symmetric, only the correlation between n(n-1) / 2 entries needs to be calculated. High correlation indicates that points that belong to the same cluster are close to each other. Not a good measure for some density or contiguity based clusters. 103
![Measuring Cluster Validity Via Correlation Chapter 8 5 7 page 551 l Correlation Measuring Cluster Validity Via Correlation [Chapter 8. 5. 7 , page 551] l Correlation](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-103.jpg)
Measuring Cluster Validity Via Correlation [Chapter 8. 5. 7 , page 551] l Correlation of incidence and proximity matrices for the K-means clusterings of the following two data sets. Corr = -0. 9235 2010/5/7 Corr = -0. 5810 104
![Statistical Framework for SSE l Chapter 8 5 8 page 554 Example Statistical Framework for SSE l [Chapter 8. 5. 8 , page 554] Example –](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-104.jpg)
Statistical Framework for SSE l [Chapter 8. 5. 8 , page 554] Example – Compare SSE of 0. 005 against three clusters in random data – Histogram shows SSE of three clusters in 500 sets of random data points of size 100 distributed over the range 0. 2 – 0. 8 for x and y values 2010/5/7 105
![Statistical Framework for Correlation Chapter 8 5 8 l Correlation of incidence and proximity Statistical Framework for Correlation [Chapter 8. 5. 8] l Correlation of incidence and proximity](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-105.jpg)
Statistical Framework for Correlation [Chapter 8. 5. 8] l Correlation of incidence and proximity matrices for the K-means clusterings of the following two data sets. Corr = -0. 9235 2010/5/7 Corr = -0. 5810 106
![Final Comment on Cluster Validity Chapter 8 5 8 The validation of clustering structures Final Comment on Cluster Validity [Chapter 8. 5. 8] “The validation of clustering structures](https://slidetodoc.com/presentation_image_h/03fe96e387ded8579cbc62b61dea1be8/image-106.jpg)
Final Comment on Cluster Validity [Chapter 8. 5. 8] “The validation of clustering structures is the most difficult and frustrating part of cluster analysis. Without a strong effort in this direction, cluster analysis will remain a black art accessible only to those true believers who have experience and great courage. ” Algorithms for Clustering Data, Jain and Dubes 2010/5/7 107