Cloning DNA Sequences that Encode Eukaryotic Protein n
- Slides: 21

Cloning DNA Sequences that Encode Eukaryotic Protein n Eukaryotic --- gene contains introns. --- m. RNA does not have introns. m. RNA c. DNA n m. RNA --- G cap at 5’ end (methylated guanine nucleotide which is joined after transcription) --- poly(A) tail at 3’ end

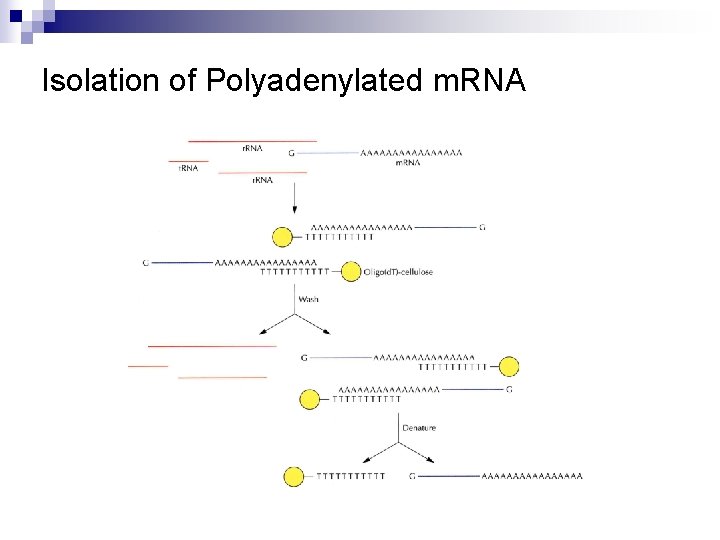
Isolation of Polyadenylated m. RNA

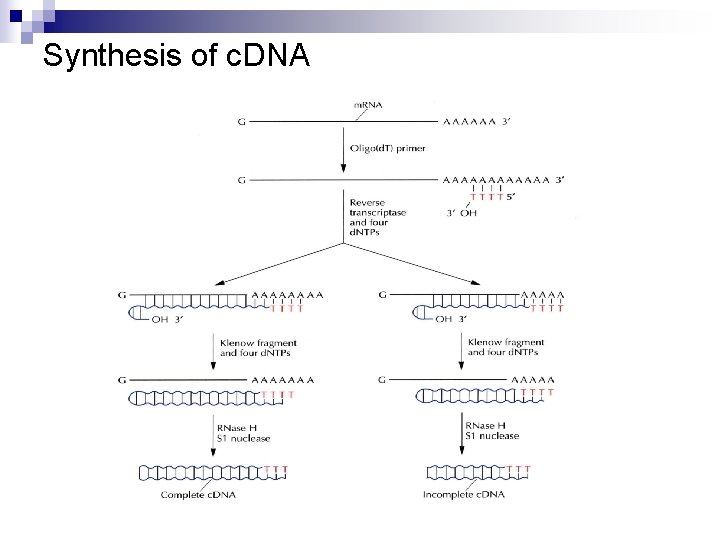
Synthesis of c. DNA

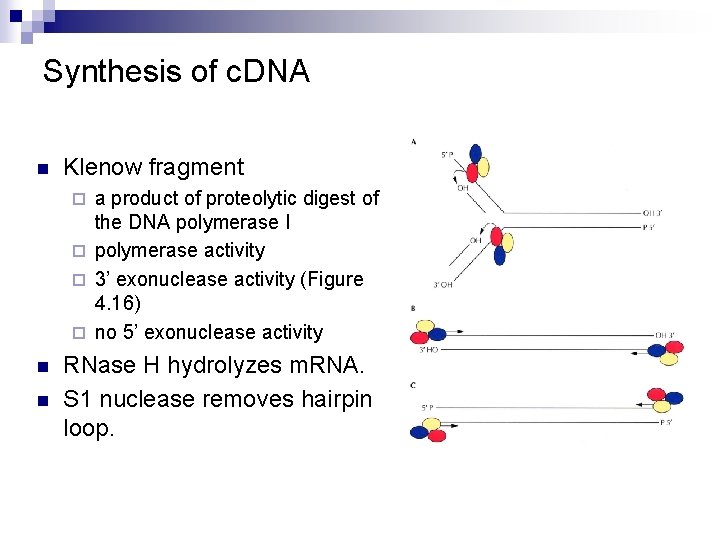
Synthesis of c. DNA n Klenow fragment a product of proteolytic digest of the DNA polymerase I ¨ polymerase activity ¨ 3’ exonuclease activity (Figure 4. 16) ¨ no 5’ exonuclease activity ¨ n n RNase H hydrolyzes m. RNA. S 1 nuclease removes hairpin loop.

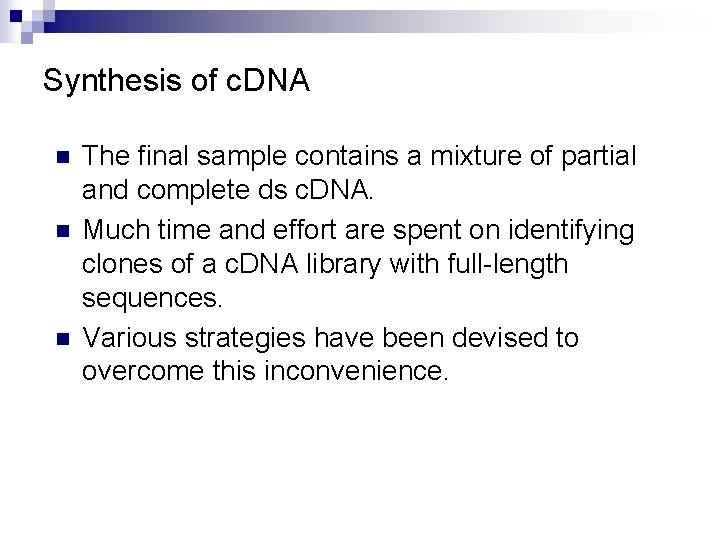
Synthesis of c. DNA n n n The final sample contains a mixture of partial and complete ds c. DNA. Much time and effort are spent on identifying clones of a c. DNA library with full-length sequences. Various strategies have been devised to overcome this inconvenience.

Cloning of Full-Length c. DNA Molecule Restriction site 5 -methyl-d. CTP (Methylation protects c. DNA from cleavage by restriction enzyme. ) Biotin Streptavidin-coated magnetic bead

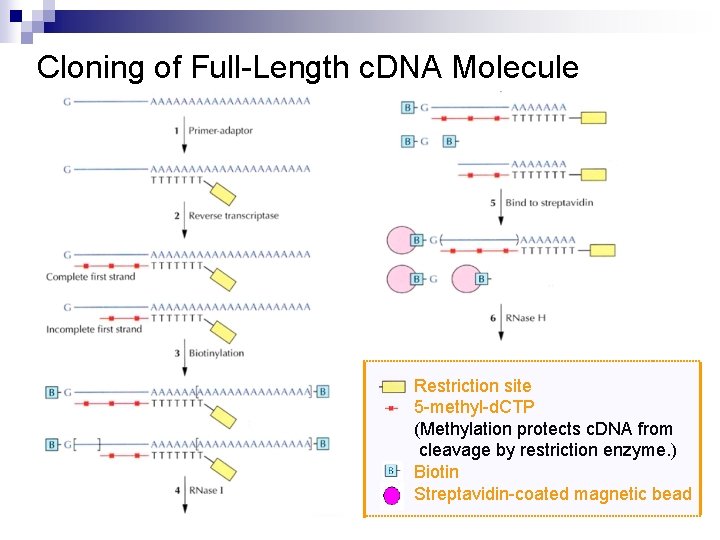
Cloning of Full-Length c. DNA Molecule : Restriction site * RNase H removes any RNA that escaped the previous treatment. * DNA ligase joins DNA segments that were synthesized internally from bits of m. RNA that escaped degradation.

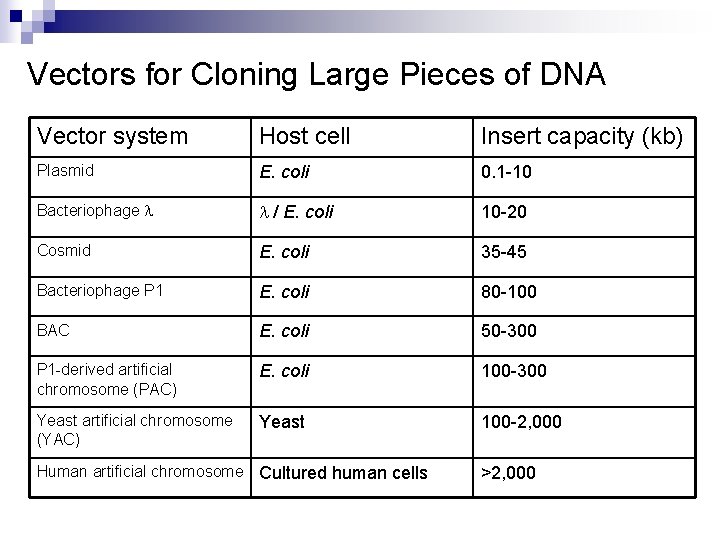
Vectors for Cloning Large Pieces of DNA Vector system Host cell Insert capacity (kb) Plasmid E. coli 0. 1 -10 Bacteriophage l l / E. coli 10 -20 Cosmid E. coli 35 -45 Bacteriophage P 1 E. coli 80 -100 BAC E. coli 50 -300 P 1 -derived artificial chromosome (PAC) E. coli 100 -300 Yeast artificial chromosome (YAC) Yeast 100 -2, 000 Human artificial chromosome Cultured human cells >2, 000

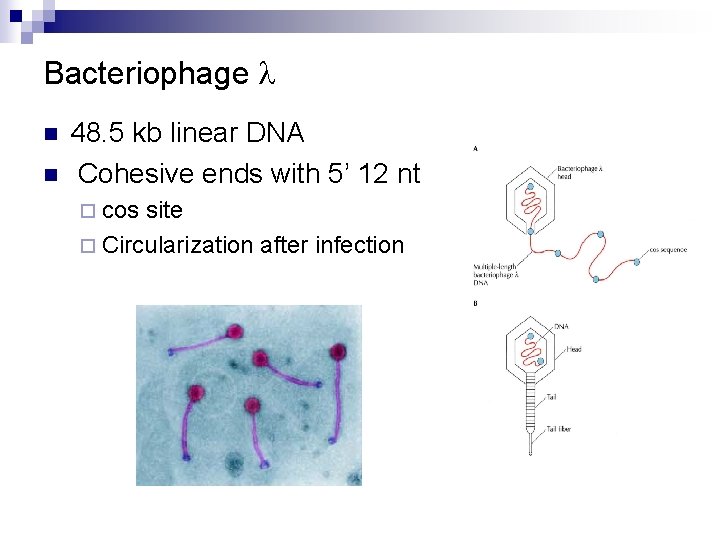
Bacteriophage l n n 48. 5 kb linear DNA Cohesive ends with 5’ 12 nt ¨ cos site ¨ Circularization after infection

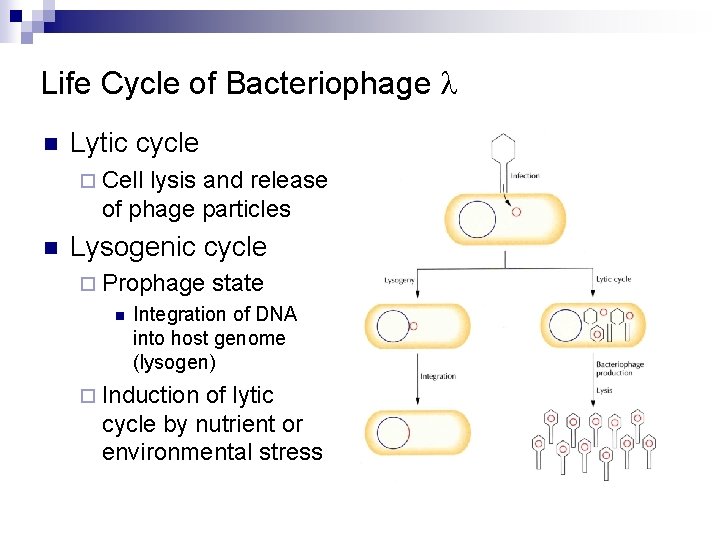
Life Cycle of Bacteriophage l n Lytic cycle ¨ Cell lysis and release of phage particles n Lysogenic cycle ¨ Prophage n state Integration of DNA into host genome (lysogen) ¨ Induction of lytic cycle by nutrient or environmental stress

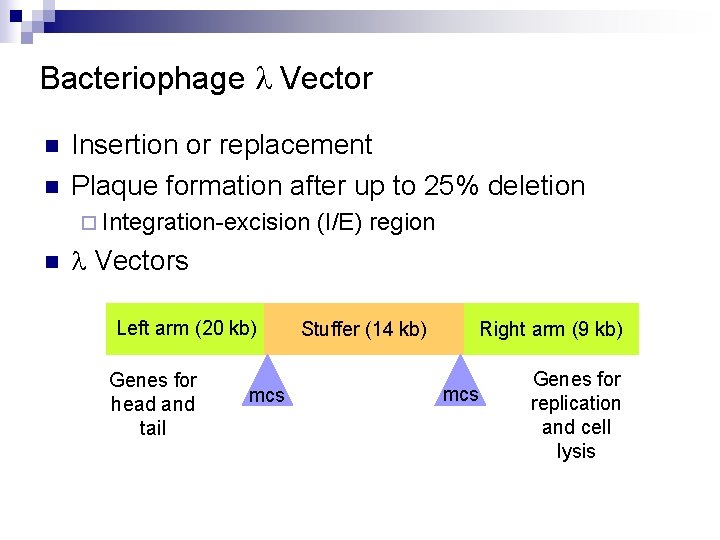
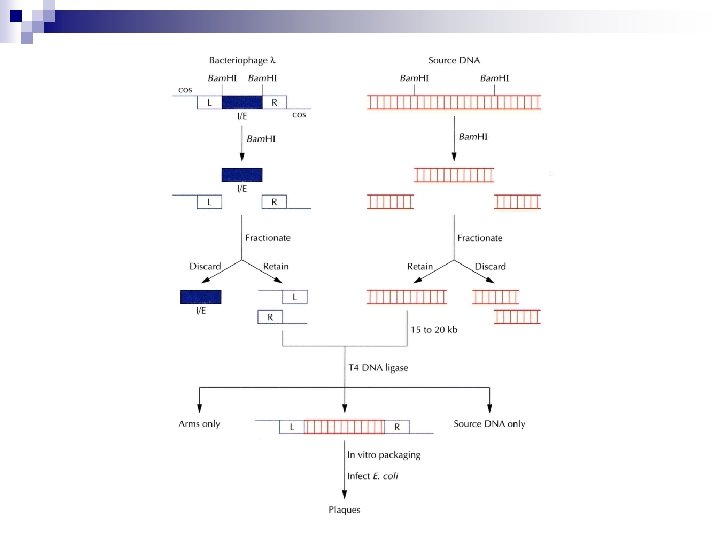
Bacteriophage l Vector n n Insertion or replacement Plaque formation after up to 25% deletion ¨ Integration-excision n (I/E) region l Vectors Left arm (20 kb) Genes for head and tail mcs Stuffer (14 kb) Right arm (9 kb) mcs Genes for replication and cell lysis


Cosmid Vector n Cosmid vector ¨ Plasmid n with l cos sites l vector vs. cosmid vector Size for packaging: 38~52 kb (75 -105% of l DNA) ¨ DNA n l Phage vectors: ¨ n Limitation for the deletion of essential genes Cosmid vectors: ¨ Accommodate 33 -47 kb DNA in 5 kb cosmid vector

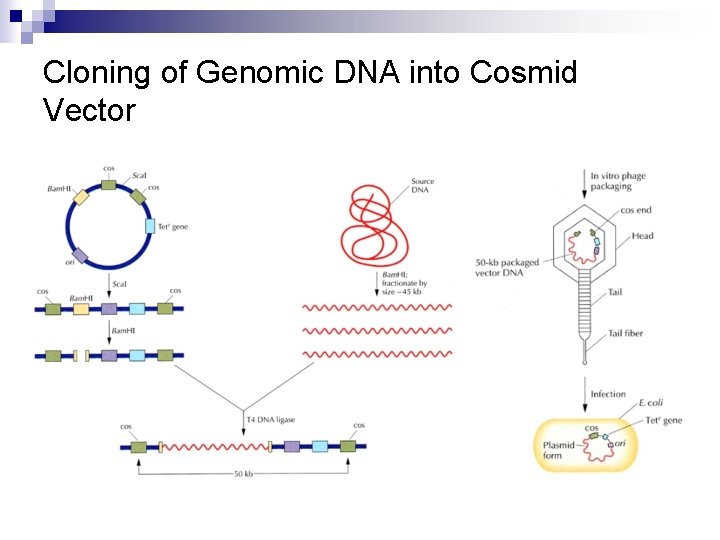
Cloning of Genomic DNA into Cosmid Vector

High-Capacity Bacterial Vector Systems n Bacteriophage P 1 -derived artificial chromosomes ¨ 100~300 n kb of insert DNA Bacterial artificial chromosome (BAC) ¨ 50~300 kb of insert DNA ¨ F-plasmid-based DNA insert-vector construct

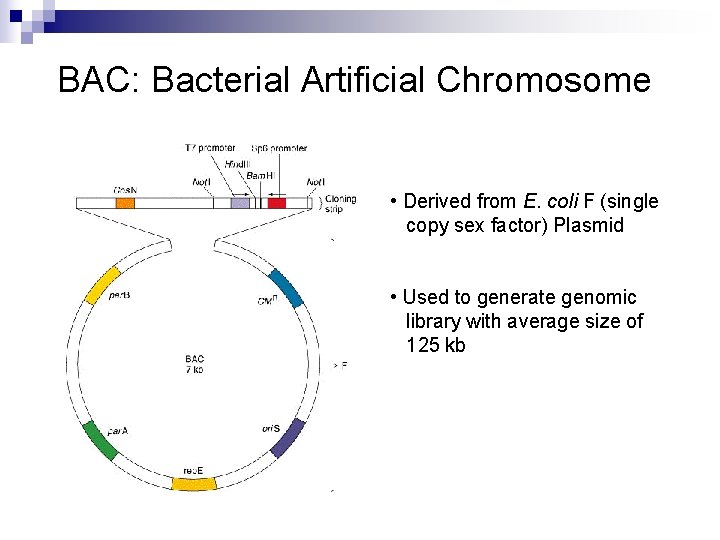
BAC: Bacterial Artificial Chromosome • Derived from E. coli F (single copy sex factor) Plasmid • Used to generate genomic library with average size of 125 kb

Genetic Transformation of Prokaryotes n Chemical method: Ca. Cl 2 and heat shock ¨ Transformation frequency n 10 -3 (one transformed cell/1000 cells) ¨ Transformation efficiency : n 107 to 108 (transformed colonies / μg of intact plasmid) ¨ Competent cells n cells that are able to take up DNA (plasmid)

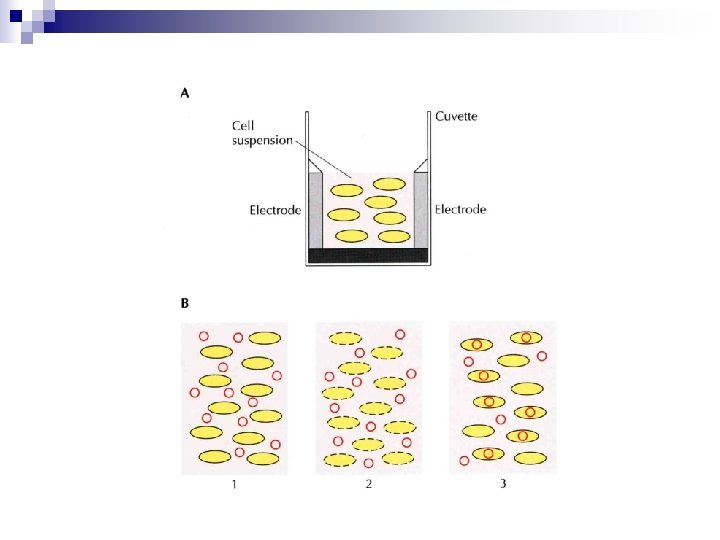
Genetic Transformation of Prokaryotes n Electroporation ¨ Electric field-mediated membrane permeabilization ¨ Effective way for large plasmids ( > 100 kb) ¨ E. coli n electric pulse of 25 m. F, 2. 5 k. V, 200 ohms for 4. 6 ms ¨ Transformation efficiency : 106 transformants (for ~136 kb) to 109 transformants (for ~3 kb) /mg DNA


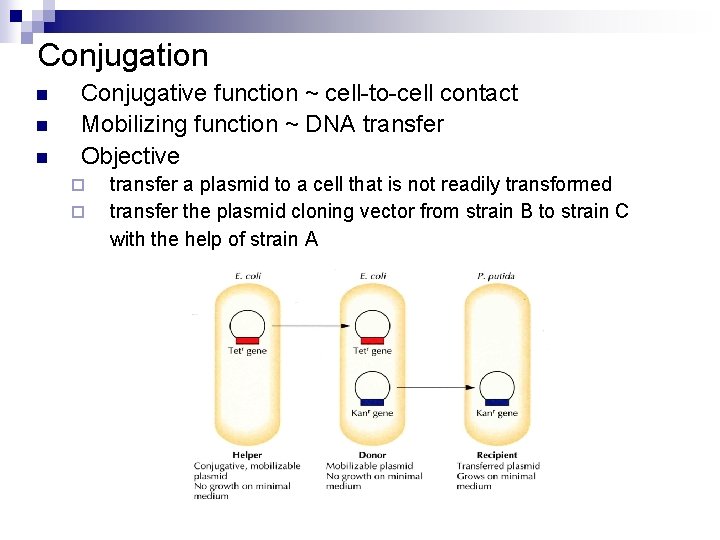
Conjugation n Conjugative function ~ cell-to-cell contact Mobilizing function ~ DNA transfer Objective ¨ ¨ transfer a plasmid to a cell that is not readily transformed transfer the plasmid cloning vector from strain B to strain C with the help of strain A

Conjugation n n Strain A contains a conjugative, mobilizing plasmid. Strain B contains a mobilizing plasmid. Strain C grows on minimal medium. Culture in complete growth medium (w/o kanamycin) for conjugation Selection in minimal medium (with kanamycin) for strain C containing the target plasmid Occasionally, the targeted recipient cell may receive both types of plasmids. These cells can be excluded by antibiotic selection.