Clin Var Using Centralized Databases to Interpret Sequence
Clin. Var: Using Centralized Databases to Interpret Sequence Variants Donna Maglott, Ph. D. Senior Staff Scientist maglott@ncbi. nlm. nih. gov www. ncbi. nlm. nih. gov. /clinvar
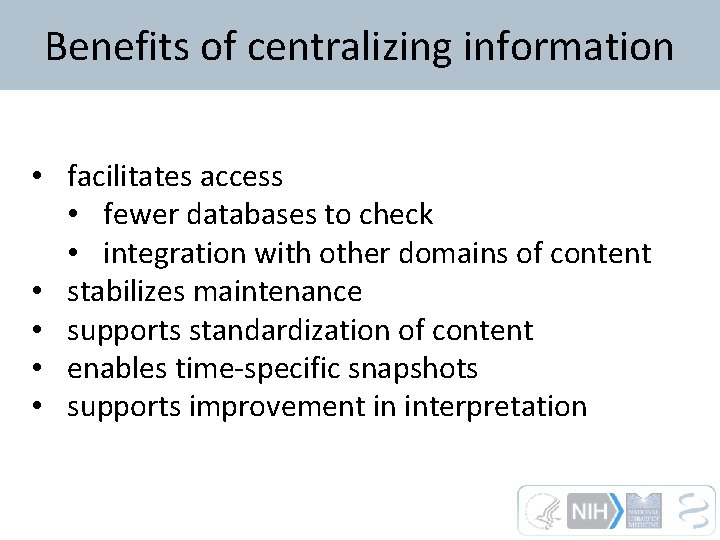
Benefits of centralizing information • facilitates access • fewer databases to check • integration with other domains of content • stabilizes maintenance • supports standardization of content • enables time-specific snapshots • supports improvement in interpretation
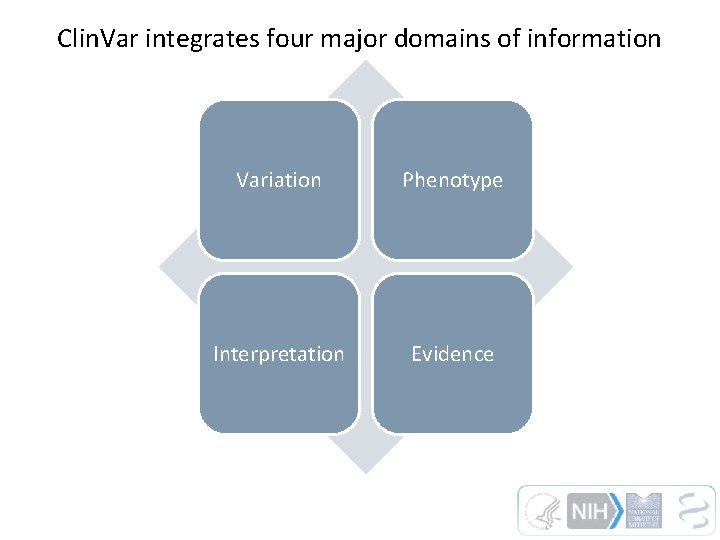
Clin. Var integrates four major domains of information Variation Phenotype Interpretation Evidence
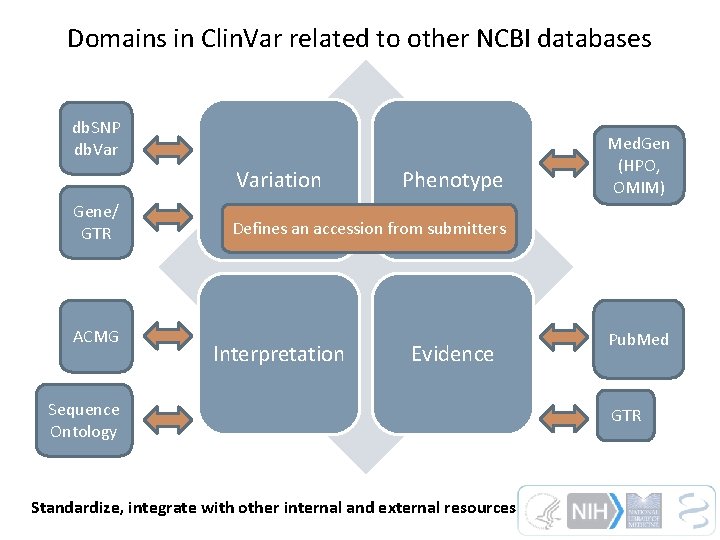
Domains in Clin. Var related to other NCBI databases db. SNP db. Variation Gene/ GTR ACMG Phenotype Med. Gen (HPO, OMIM) Defines an accession from submitters Interpretation Evidence Sequence Ontology Standardize, integrate with other internal and external resources Pub. Med GTR
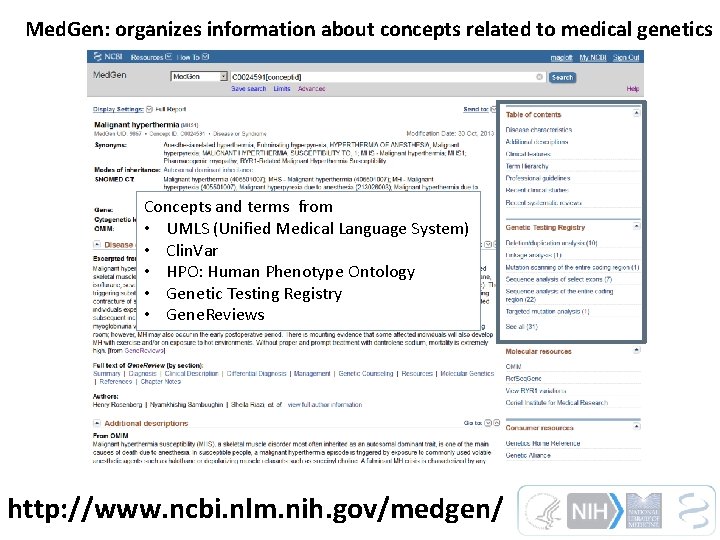
Med. Gen: organizes information about concepts related to medical genetics Concepts and terms from • UMLS (Unified Medical Language System) • Clin. Var • HPO: Human Phenotype Ontology • Genetic Testing Registry • Gene. Reviews http: //www. ncbi. nlm. nih. gov/medgen/
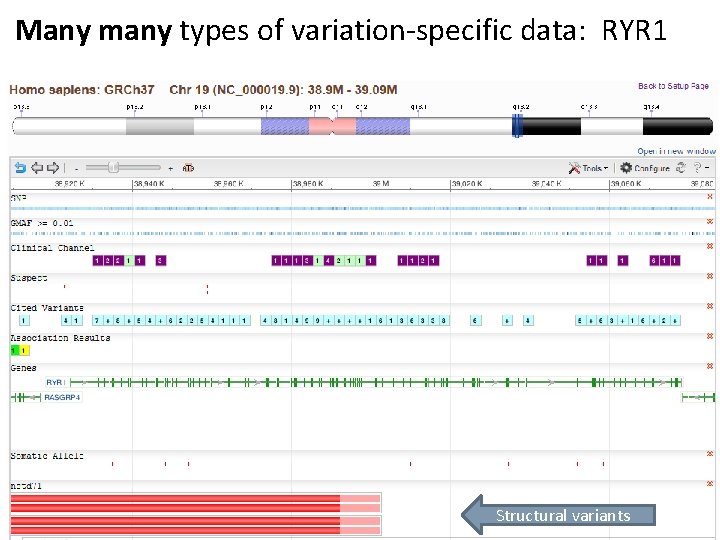
Many many types of variation-specific data: RYR 1 Structural variants

Clin. Gen: Making sense of all the data http: //www. nih. gov/news/health/sep 2013/nhgri-25. htm

Clin. Gen and Clin. Var

What is Clin. Var? http: //www. ncbi. nlm. nih. gov/clinvar/
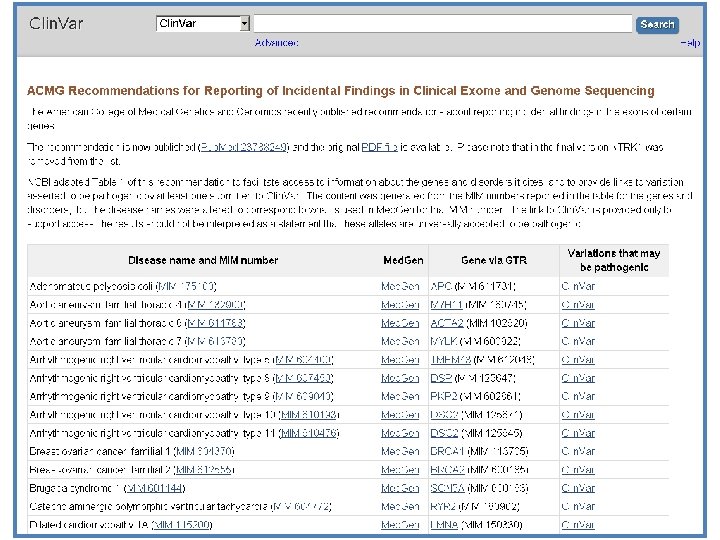
Clin. Var has multiple functions • Standardize representation of alleles, phenotypes, and interpretation • Archive submitted interpretations of variation/phenotype relationships (i. e. keep history and support re-interpretation) • Report whether interpretations are consistent or conflicting interpretations • Disseminate content via web, ftp, or API • Provide attribution to submitters
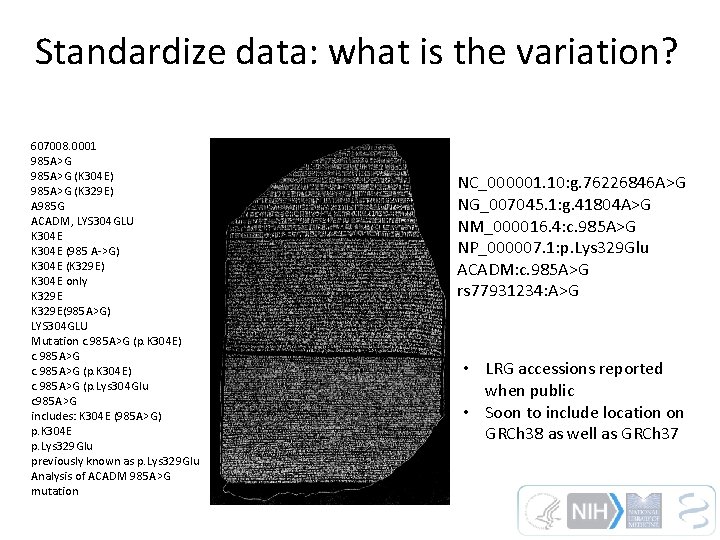
Standardize data: what is the variation? 607008. 0001 985 A>G (K 304 E) 985 A>G (K 329 E) A 985 G ACADM, LYS 304 GLU K 304 E (985 A->G) K 304 E (K 329 E) K 304 E only K 329 E(985 A>G) LYS 304 GLU Mutation c. 985 A>G (p. K 304 E) c. 985 A>G (p. Lys 304 Glu c 985 A>G includes: K 304 E (985 A>G) p. K 304 E p. Lys 329 Glu previously known as p. Lys 329 Glu Analysis of ACADM 985 A>G mutation NC_000001. 10: g. 76226846 A>G NG_007045. 1: g. 41804 A>G NM_000016. 4: c. 985 A>G NP_000007. 1: p. Lys 329 Glu ACADM: c. 985 A>G rs 77931234: A>G • LRG accessions reported when public • Soon to include location on GRCh 38 as well as GRCh 37
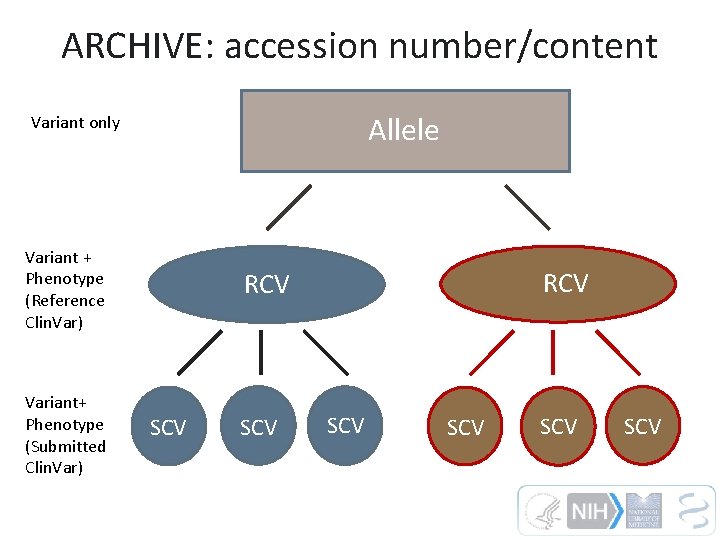
ARCHIVE: accession number/content Allele Variant only Variant + Phenotype (Reference Clin. Var) Variant+ Phenotype (Submitted Clin. Var) RCV SCV SCV SCV
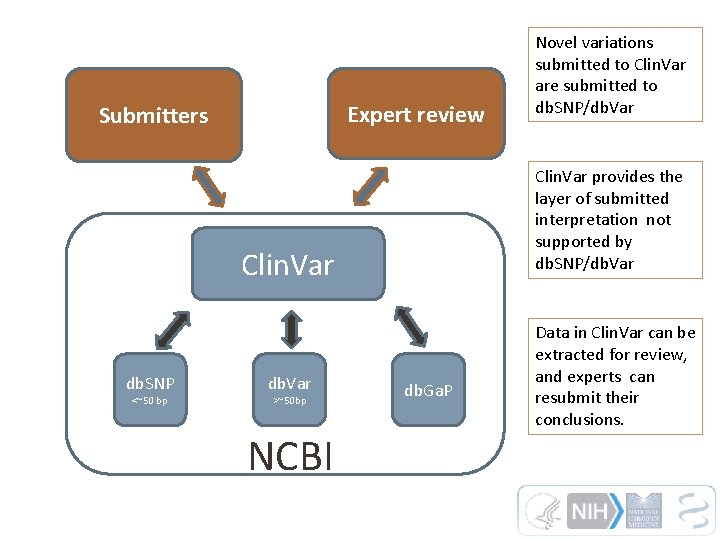
Expert review Submitters Clin. Var provides the layer of submitted interpretation not supported by db. SNP/db. Var Clin. Var db. SNP <~50 bp db. Var >~50 bp NCBI Novel variations submitted to Clin. Var are submitted to db. SNP/db. Var db. Ga. P Data in Clin. Var can be extracted for review, and experts can resubmit their conclusions.
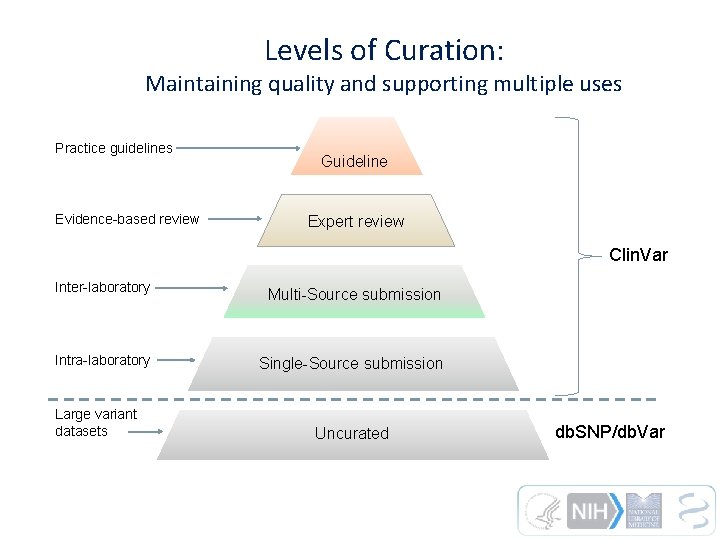
Levels of Curation: Maintaining quality and supporting multiple uses Practice guidelines Evidence-based review Guideline Expert review Clin. Var Inter-laboratory Multi-Source submission Intra-laboratory Single-Source submission Large variant datasets Uncurated db. SNP/db. Var
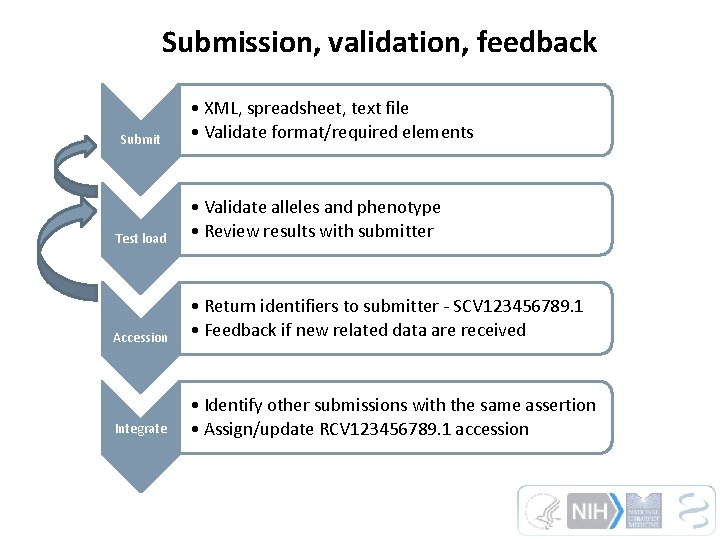
Submission, validation, feedback Submit • XML, spreadsheet, text file • Validate format/required elements Test load • Validate alleles and phenotype • Review results with submitter Accession • Return identifiers to submitter - SCV 123456789. 1 • Feedback if new related data are received Integrate • Identify other submissions with the same assertion • Assign/update RCV 123456789. 1 accession
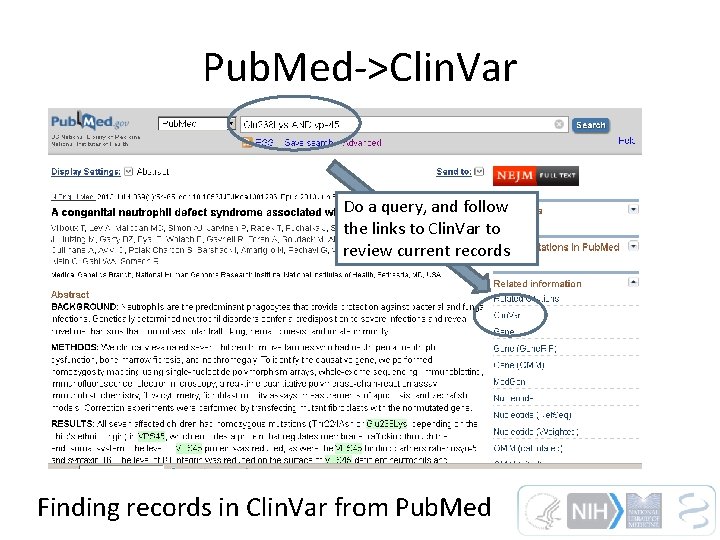
Pub. Med->Clin. Var Do a query, and follow the links to Clin. Var to review current records Finding records in Clin. Var from Pub. Med
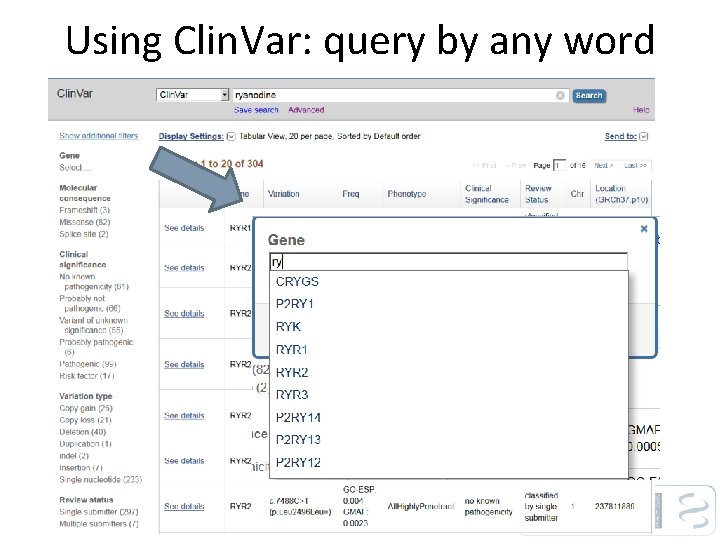
Using Clin. Var: query by any word
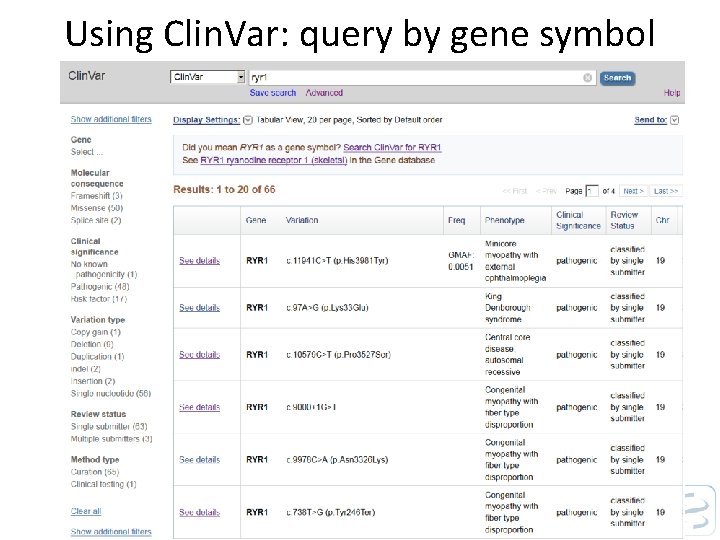
Using Clin. Var: query by gene symbol
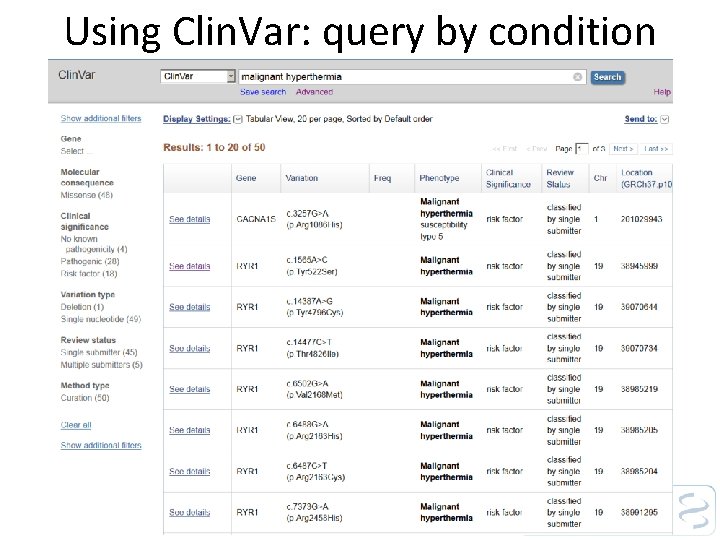
Using Clin. Var: query by condition
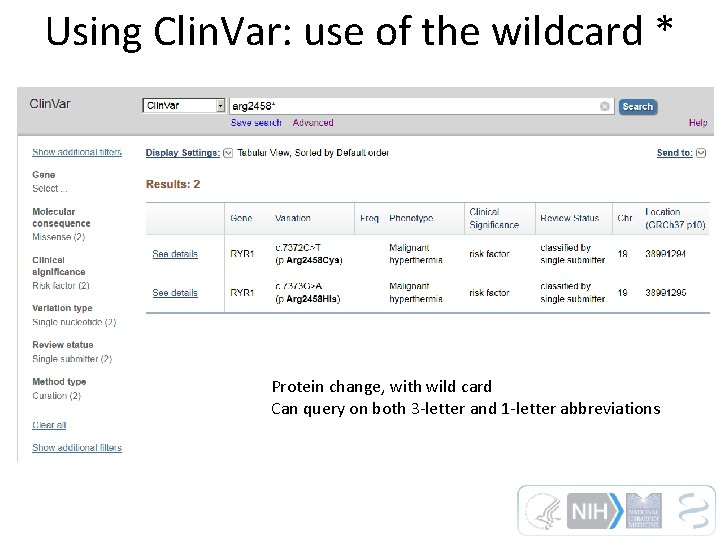
Using Clin. Var: use of the wildcard * Protein change, with wild card Can query on both 3 -letter and 1 -letter abbreviations
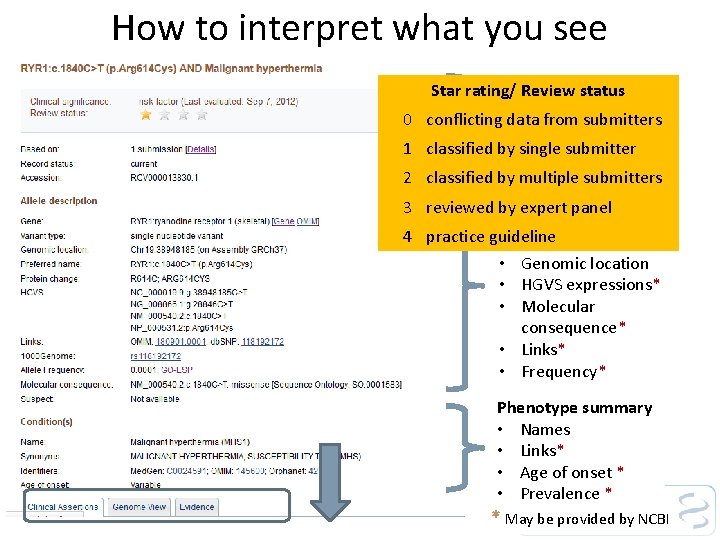
How to interpret what you see 0 1 2 3 4 Star rating/ Review status Interpretation Significance (dated) conflicting • data from submitters • Review status * classified by submitter * • single Accession. version classified by multiple submitters Allele summary reviewed by expert panel • Gene practice guideline • Variant type • Genomic location • HGVS expressions* • Molecular consequence* • Links* • Frequency* Phenotype summary • Names • Links* • Age of onset * • Prevalence * * May be provided by NCBI
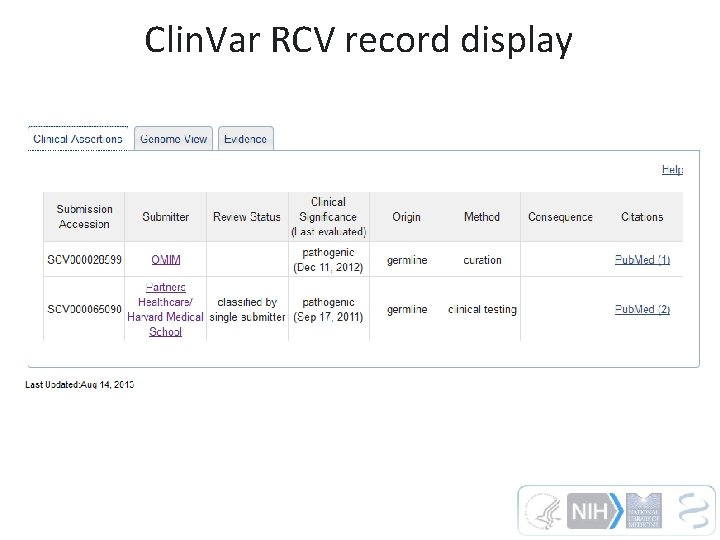
Clin. Var RCV record display
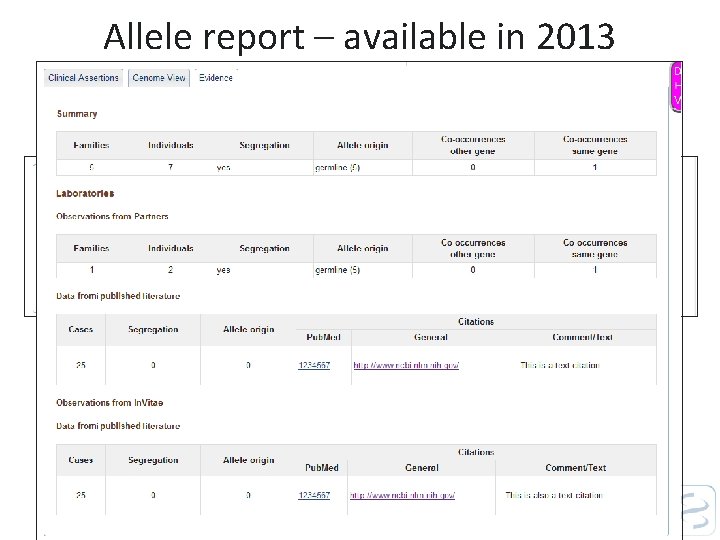
Allele report – available in 2013
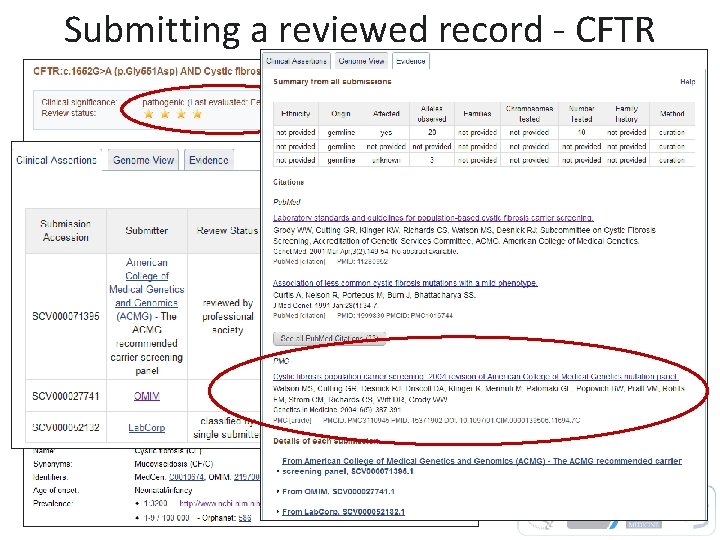
Submitting a reviewed record - CFTR

Acknowledgements Clin. Var/Gene/GTR/ Med. Gen/Ref. Seq. Gene Alex Astashyn Shanmuga Chitipiralla Viatcheslav Gorelenkov Baoshan Gu Douglas Hoffman Wonhee Jang Brandi Kattman Ken Katz Melissa Landrum Jennifer Lee Adriana Malheiro Michael Ovetsky George Riley Wendy Rubinstein Amanjeev Sethi Ray Tully Ricardo Villamarin db. SNP/db. Var/db. Ga. P ICCG/Clin. Gen Deanna Church Michael Feolo Lon Phan Ming Ward John Garner Tim Hefferon Brad Holmes John Lopez Rama Maiti Jose Mena David Shao Sherri Bale Madhuri Hegde Christa Martin Joyce Mitchell Heidi Rehm Erin Riggs …and many others All of NCBI Jim Ostell Steve Sherry Submitters SCRP …http: //www. ncbi. nlm. nih. gov/clinvar/submitters/

Submissions welcomed! http: //www. ncbi. nlm. nih. gov/clinvar/docs/submit/ clinvar@ncbi. nlm. nih. gov info@ncbi. nlm. nih. gov The accuracy and completeness of the databases depend on submissions and feedback
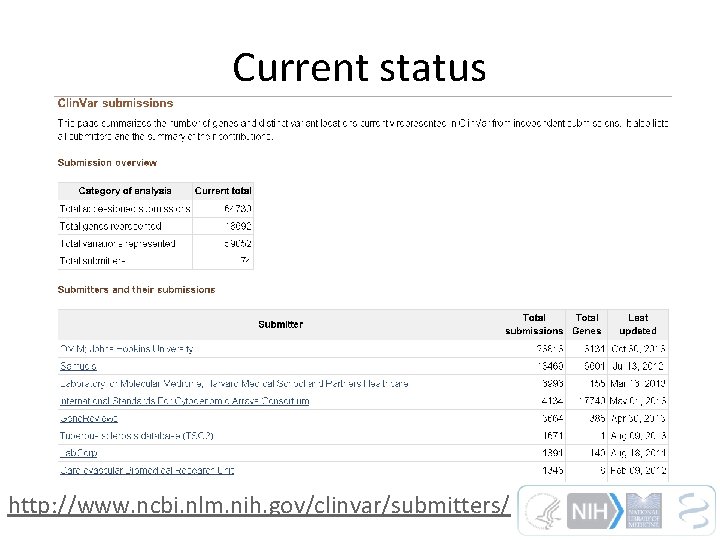
Current status http: //www. ncbi. nlm. nih. gov/clinvar/submitters/
- Slides: 28