Classifying MSA Packages Multiple Sequence Alignments in the

















- Slides: 58

Classifying MSA Packages Multiple Sequence Alignments in the Genome Era Cédric Notredame Information Génétique et Structurale CNRS-Marseille, France

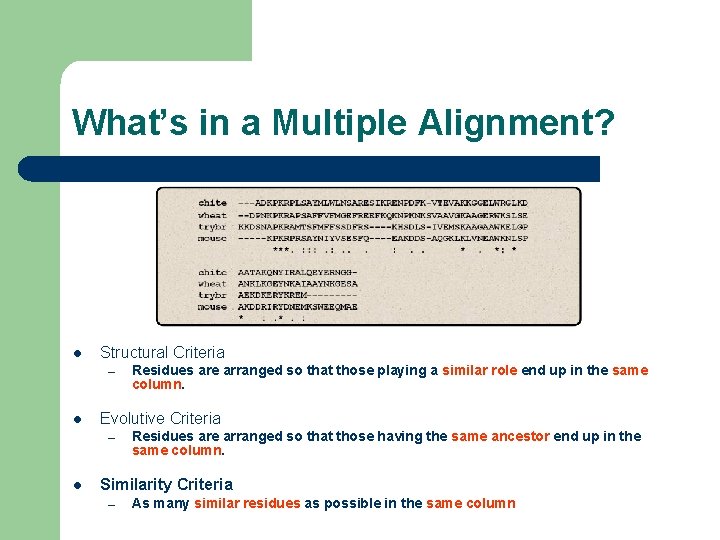
What’s in a Multiple Alignment? l Structural Criteria – l Evolutive Criteria – l Residues are arranged so that those playing a similar role end up in the same column. Residues are arranged so that those having the same ancestor end up in the same column. Similarity Criteria – As many similar residues as possible in the same column

What’s in a Multiple Alignment?

What’s in a Multiple Alignment? l l The MSA contains what you put inside… You can view your MSA as: – – – A record of evolution A summary of a protein family A collection of experiments made for you by Nature…

What’s in a Multiple Alignment?

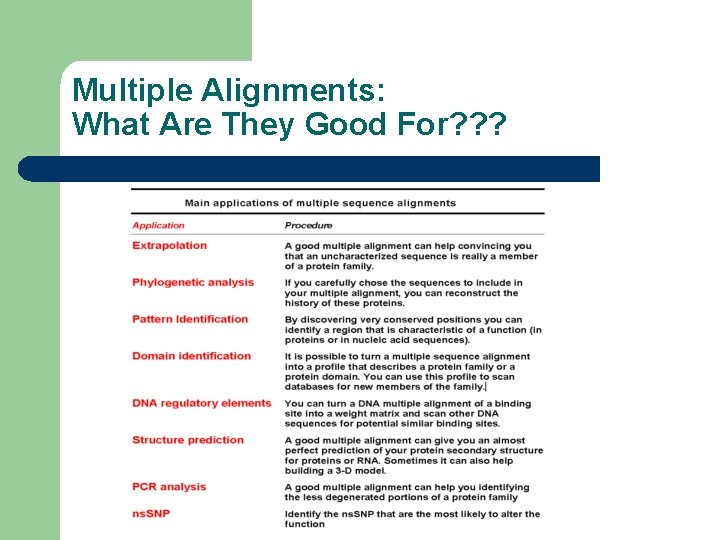
Multiple Alignments: What Are They Good For? ? ?

Computing the Correct Alignement is a Complicated Problem

A Taxonomy of Multiple Sequence Alignment Packages Objective Function Assembly Algorithms

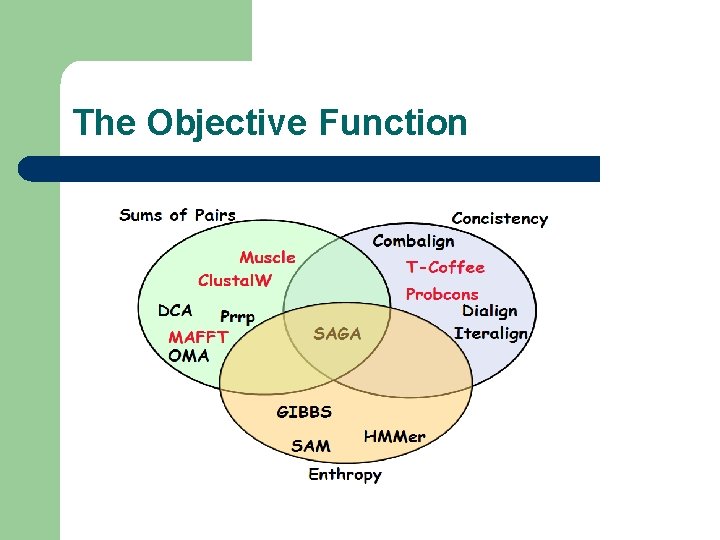
The Objective Function

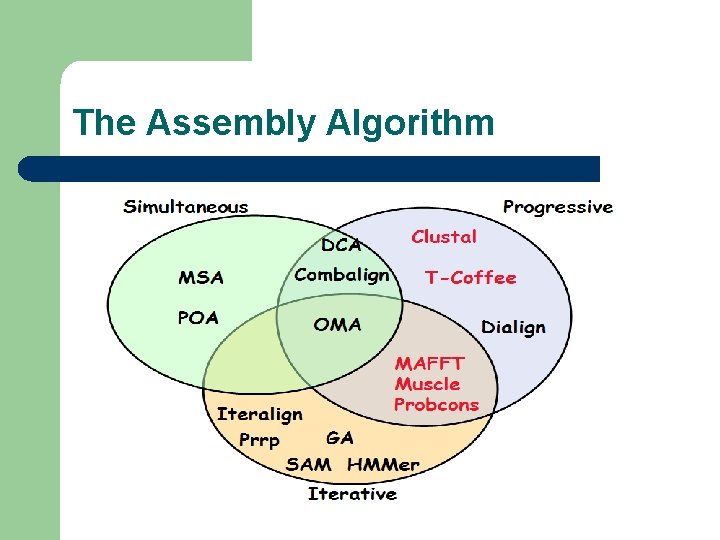
The Assembly Algorithm

A Tale of Three Algorithms l Progressive: Clustal. W l Iterative: Muscle l Concistency Based: T-Coffee and Probcons

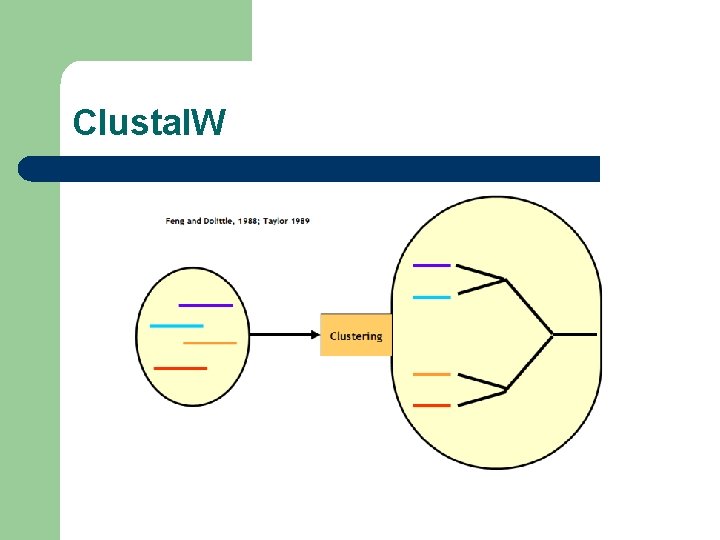
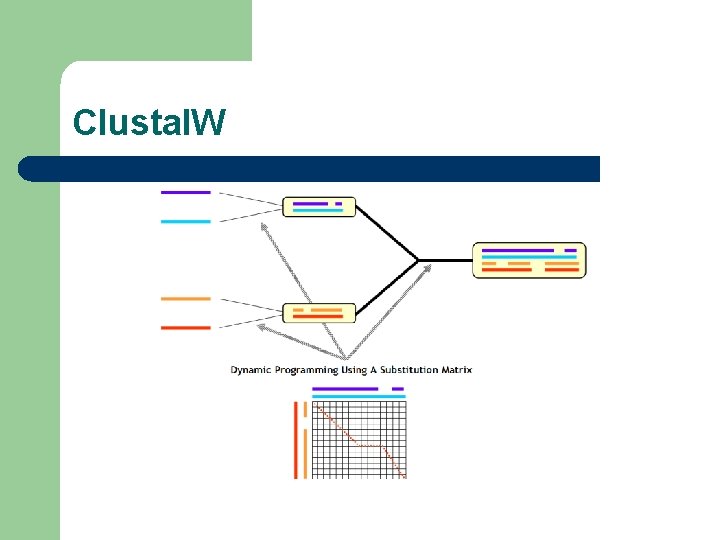
Clustal. W Algorithm l l l Paula Hogeweg: First Description (1981) Taylor, Dolittle: Reinvention in 1989 Higgins: Most Successful Implementation

Clustal. W

Clustal. W

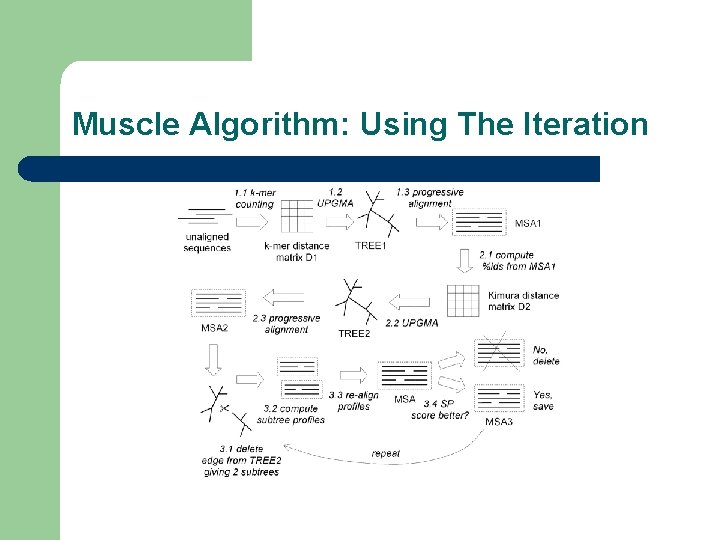
Muscle Algorithm: Using The Iteration l AMPS: First iterative Algorithm (Barton, 1987) l Stochastic methods: Genetic Algorithms and Simulated Annealing (Notredame, 1995) l Prrp: Ancestor of MUSCLE and MAFT (1996) l Muscle: the most succesful iterative strategy to this day

Muscle Algorithm: Using The Iteration

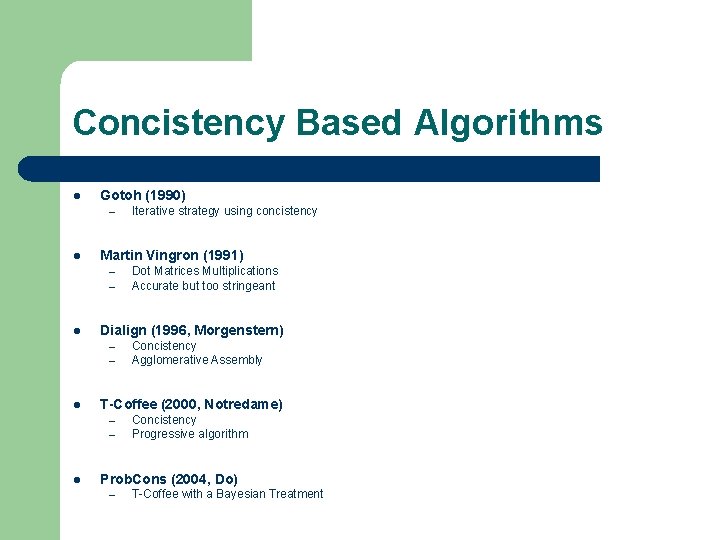
Concistency Based Algorithms l Gotoh (1990) – l Martin Vingron (1991) – – l – Concistency Agglomerative Assembly T-Coffee (2000, Notredame) – – l Dot Matrices Multiplications Accurate but too stringeant Dialign (1996, Morgenstern) – l Iterative strategy using concistency Concistency Progressive algorithm Prob. Cons (2004, Do) – T-Coffee with a Bayesian Treatment

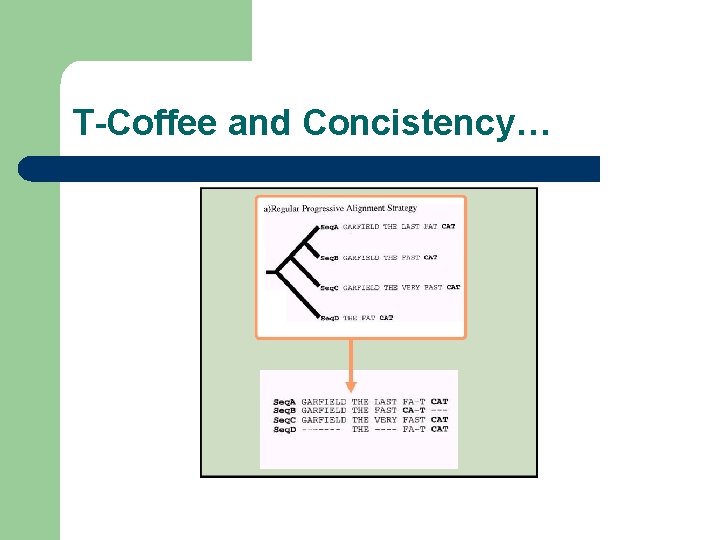
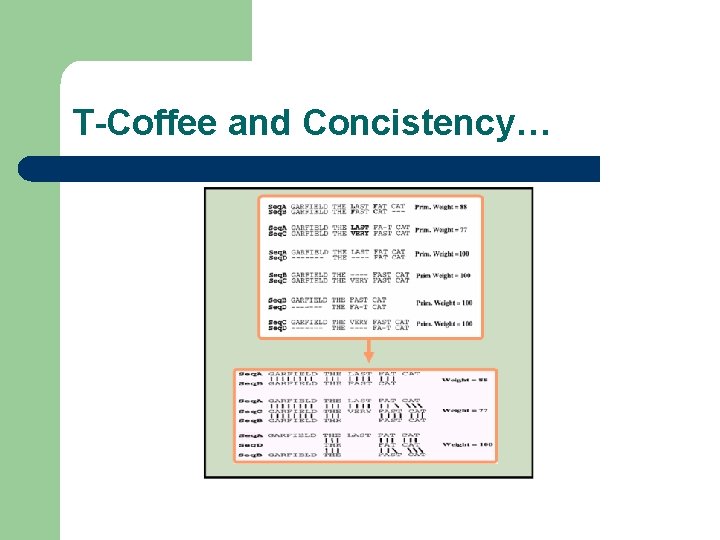
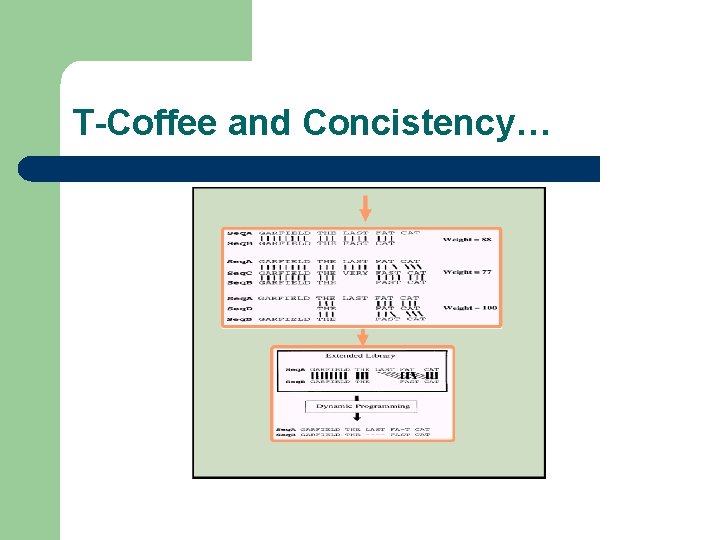
T-Coffee and Concistency…

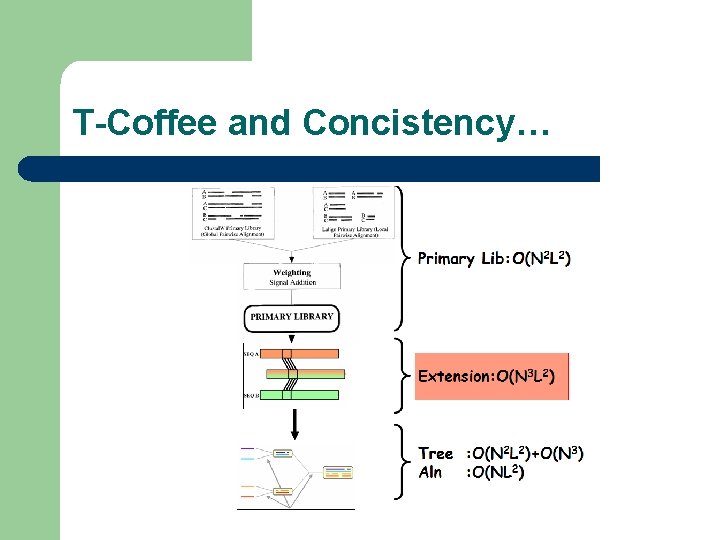
T-Coffee and Concistency…

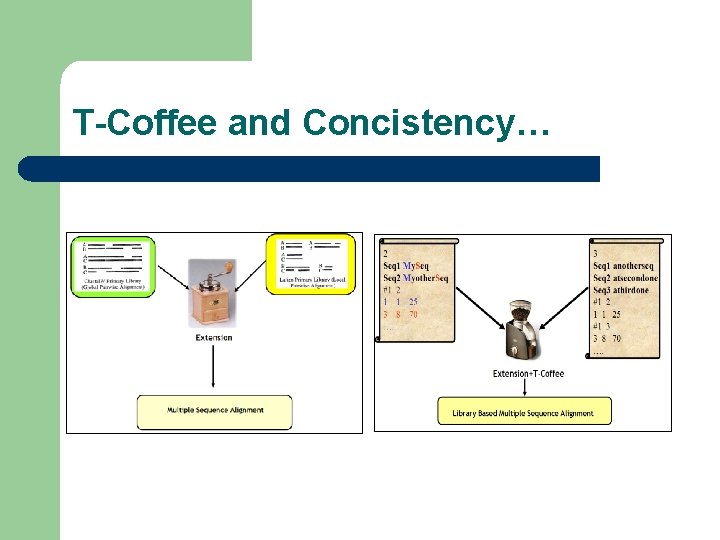
T-Coffee and Concistency…

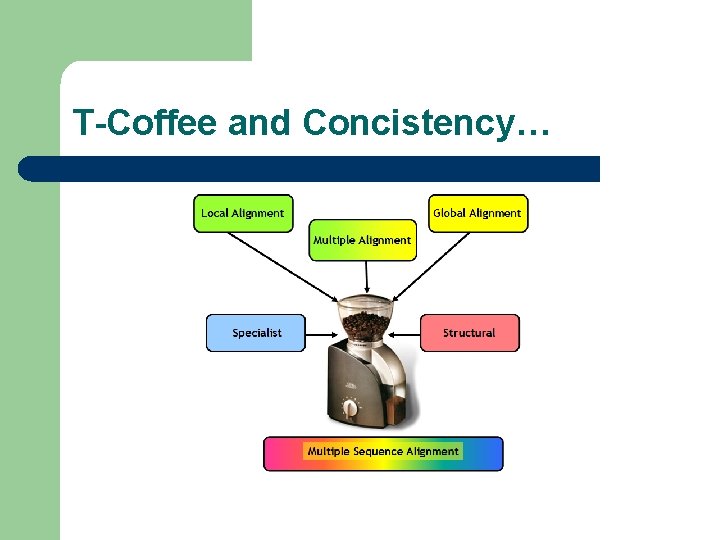
T-Coffee and Concistency…

T-Coffee and Concistency…

T-Coffee and Concistency…

T-Coffee and Concistency…

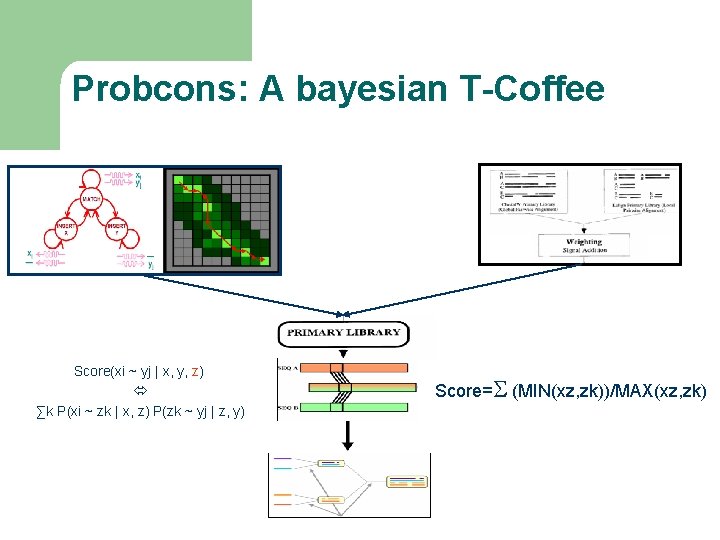
Probcons: A bayesian T-Coffee Score(xi ~ yj | x, y, z) ∑k P(xi ~ zk | x, z) P(zk ~ yj | z, y) Score=S (MIN(xz, zk))/MAX(xz, zk)

Evaluating Methods… Who is the best? Says who…?

Structures Vs Sequences

Evaluating Alignments Quality: Collections and Results

Evaluating Alignments Quality Collections l Homstrad: The most Ancient l SAB: Yet Another Benchmark l Prefab: The most extensive and automated l Bali. Base: the first designed for MSA benchmarks (Recently updated)

Homstrad (Mizuguchi, Blundell, Overington, 1998) l Hand Curated Structure Superposition Hom +0 l Not designed for Multiple Alignments Hom +3 l Biased with Clustal. W Hom +8 l No CORE annotation

Homstrad: Known issues Thiored. aln 1 aaza 1 ego 1 thx 2 trxa 3 trx 3 grx ------------ mfkvygydsnihkcvycdnakrlltvkk-----qpf ------------ mqtvifgrs----gcpycvrakdlaeklsnerddfqy skgviti-tdaefesevlkae-qpvlvyfwaswcgp cqlmsplinlaantys---drlkv sdkiihl-tddsfdtdvlkad-gailvdfwaewcgp ckmiapildeiadeyq---gkltv --mvkqiesktafqealdaagdklvvvdfsatwcgp ckmikpffhslsekys----nvif ------------ anveiytke----tcpyshrakallsskg-----vsf : . 1 aaza 1 ego 1 thx 2 trxa 3 trx 3 grx efinimpekgvfddekiaelltklgrdtqigltmpqvfapd----gshigg---fdqlre qyvdirae-----gitkedlqqkagkp---vetvpqifv-d----qqhigg---ytdfaa vkleid-----pnpttvkkykve-----gvpalrlvkgeqildstegviskdklls aklnid-----qnpgtapkygir-----giptlllfkngevaatkvgalskgqlke levdvd-----dcqdvasecevk-----ctptfqffkkgqkvgefsgan-keklea qelpidgn-----aakreemikrsgr-----ttvpqifi-d----aqhigg---yddlya : : . *. :

Homstrad

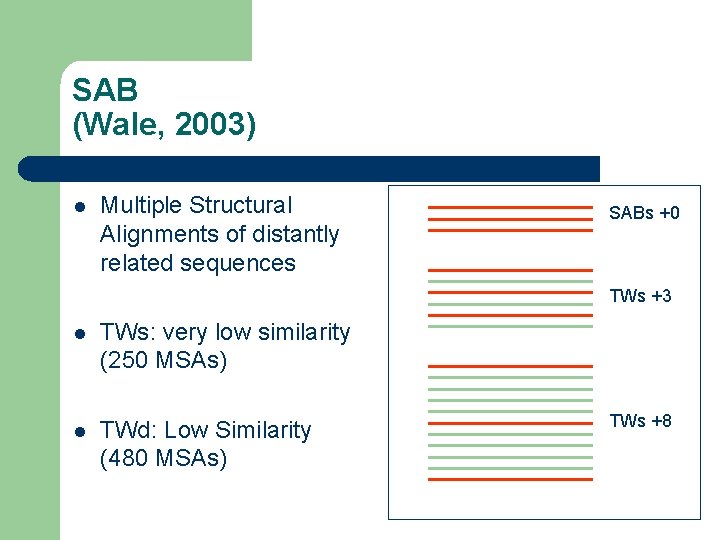
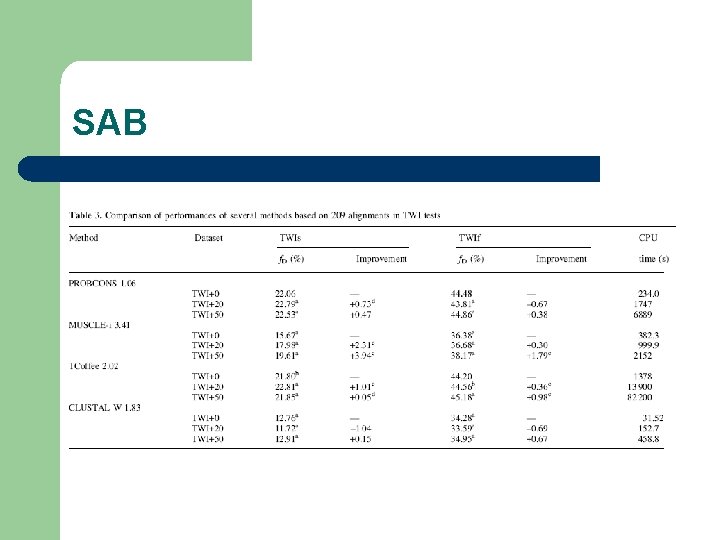
SAB (Wale, 2003) l Multiple Structural Alignments of distantly related sequences SABs +0 TWs +3 l TWs: very low similarity (250 MSAs) l TWd: Low Similarity (480 MSAs) TWs +8

SAB

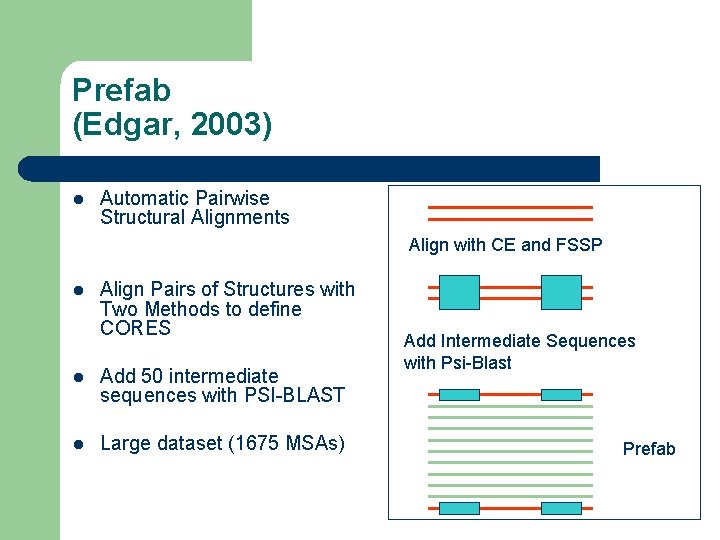
Prefab (Edgar, 2003) l Automatic Pairwise Structural Alignments Align with CE and FSSP l Align Pairs of Structures with Two Methods to define CORES l Add 50 intermediate sequences with PSI-BLAST l Large dataset (1675 MSAs) Add Intermediate Sequences with Psi-Blast Prefab

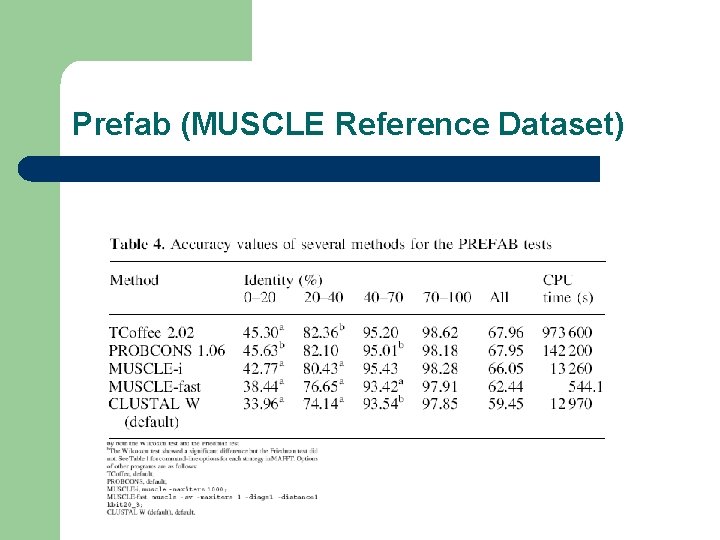
Prefab (MUSCLE Reference Dataset)

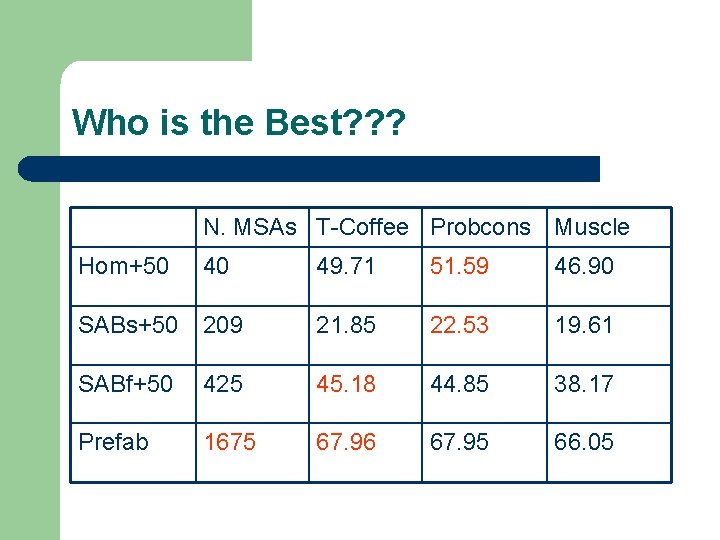
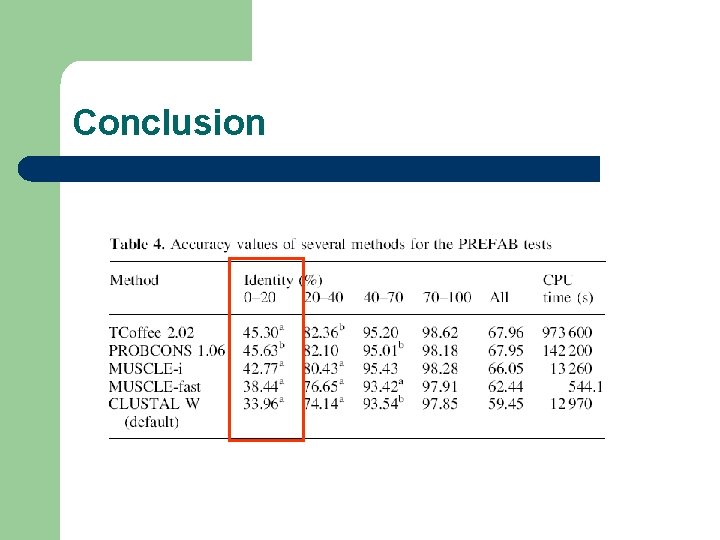
Who is the Best? ? ? N. MSAs T-Coffee Probcons Muscle Hom+50 40 49. 71 51. 59 46. 90 SABs+50 209 21. 85 22. 53 19. 61 SABf+50 425 45. 18 44. 85 38. 17 Prefab 1675 67. 96 67. 95 66. 05

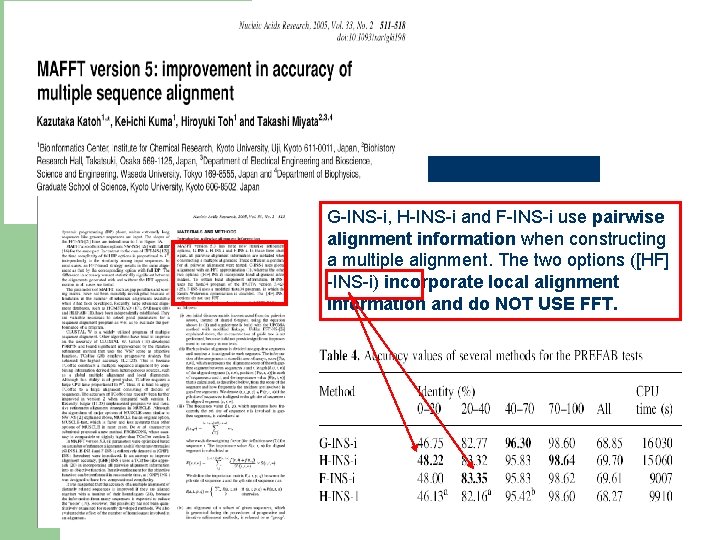
A Case for reading papers The FFT of MAFFT


G-INS-i, H-INS-i and F-INS-i use pairwise alignment information when constructing a multiple alignment. The two options ([HF] -INS-i) incorporate local alignment information and do NOT USE FFT.

Improving T-Coffee l Ease The Use Heterogenous Information – l 3 DCoffee Speed up the algorithm – – T-Coffee. DPA (Double Progressive Algorithm) Parallel T-Coffee (collaboration with EPFL)

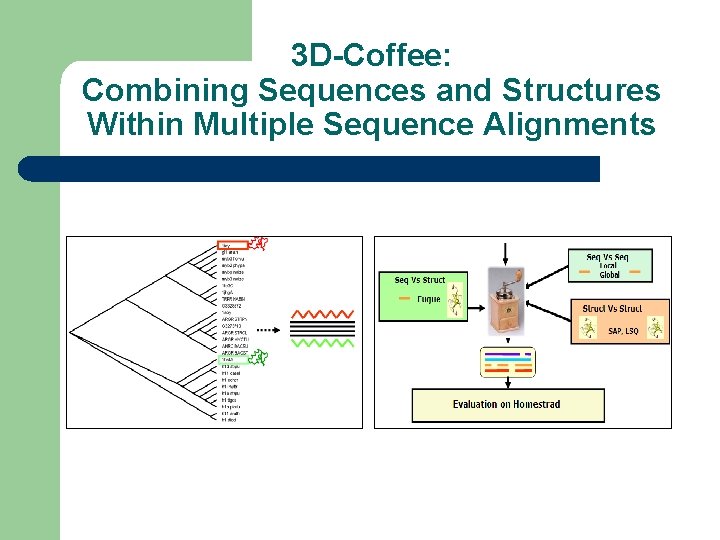
3 D-Coffee: Combining Sequences and Structures Within Multiple Sequence Alignments

3 D-Coffee: Combining Sequences and Structures Within Multiple Sequence Alignments

T-Coffee-DPA DPA: Double Progressive ALN Target: 1000 -10. 000 seq Principle: DC Progressive ALN Application: Decreasing Redundancy

Who is the Best ? ? ? l Most Packages claim to be more accurate than T-Coffee, few really are… l None of the existing packages is concistently the best: The PERFECT method does not exist

Conclusion l Concistency Based Methods Have an Edge over Conventional – – l Hard to tell Methods Appart – – l Better management of the data Better extension possibilities Reference databases are not very precise Algorithms evolve quickly Sequence Alignment is NOT a solved problem – Will be solved when Structure Prediction is solved

Conclusion

http: //igs-server. cnrs-mrs. fr/Tcoffee l l l l Fabrice Armougom Sebastien Moretti Olivier Poirot Karsten Sure Chantal Abergel Des Higgins Orla O’Sullivan Iain Wallace cedric. notredame@europe. com

Dissemination: The right Vector Amazon. com: 12/11/05 Barnes&Noble (US): 12/11/05 Amazon. co. uk: 12/11/05

Cadrie Notredom et Michael Claverie


T-Coffee-DPA l T-Coffee-DPA is about 20 times faster than the Standard T-Coffee l Preliminary tests indicate a slightly higher accuracy l Beta-Test versions will be available by September but can will be sent on request.

3 D TCoffee. DPA Vs The Human Kinome… l 521 sequences l 46 structures having 80% or more sequence identity with other kinome structures l Use of 3 D-Coffee. DPA (unpublished) developped especially for the kinome analysis

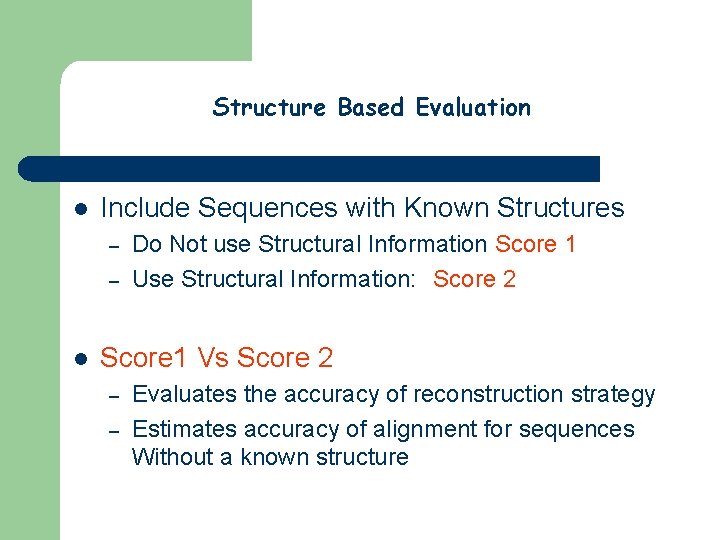
Structure Based Evaluation l Include Sequences with Known Structures – – l Do Not use Structural Information Score 1 Use Structural Information: Score 2 Score 1 Vs Score 2 – – Evaluates the accuracy of reconstruction strategy Estimates accuracy of alignment for sequences Without a known structure

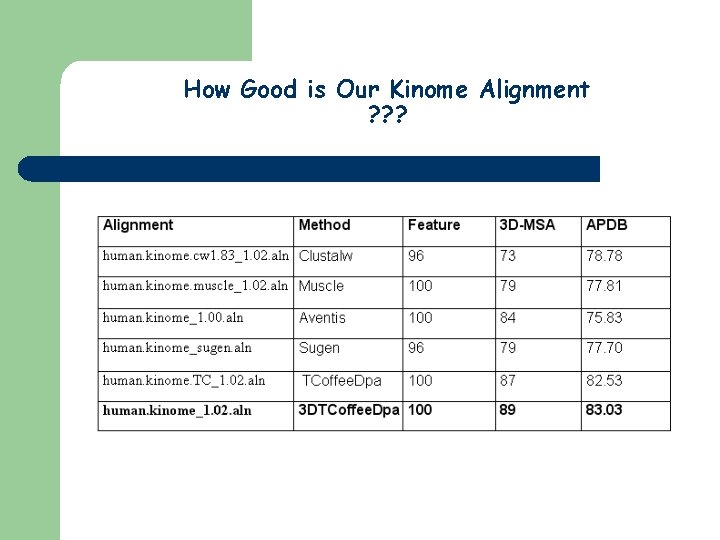
How Good is Our Kinome Alignment ? ? ?

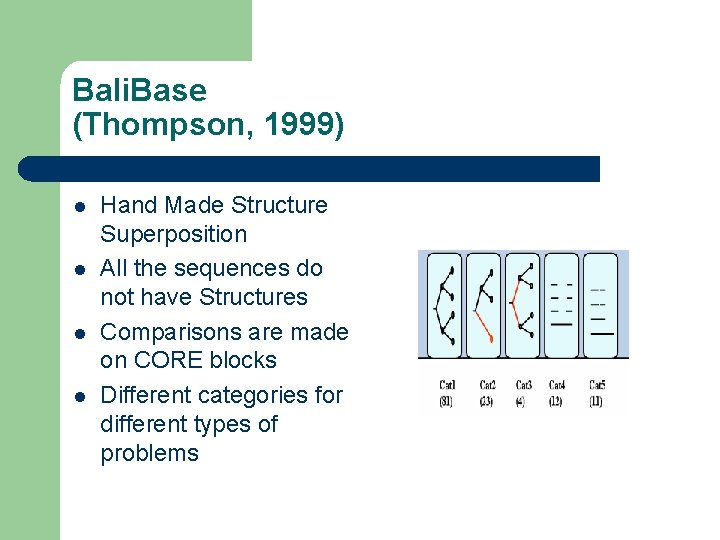
Bali. Base (Thompson, 1999) l l Hand Made Structure Superposition All the sequences do not have Structures Comparisons are made on CORE blocks Different categories for different types of problems

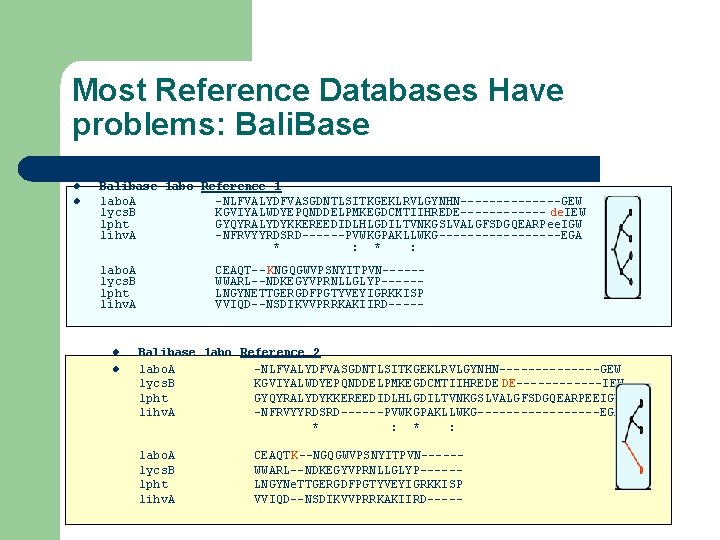
Most Reference Databases Have problems: Bali. Base l l Balibase 1 abo Reference 1 1 abo. A -NLFVALYDFVASGDNTLSITKGEKLRVLGYNHN-------GEW 1 ycs. B KGVIYALWDYEPQNDDELPMKEGDCMTIIHREDE------ de. IEW 1 pht GYQYRALYDYKKEREEDIDLHLGDILTVNKGSLVALGFSDGQEARPee. IGW 1 ihv. A -NFRVYYRDSRD------PVWKGPAKLLWKG---------EGA * : 1 abo. A 1 ycs. B 1 pht 1 ihv. A l l CEAQT--KNGQGWVPSNYITPVN-----WWARL--NDKEGYVPRNLLGLYP-----LNGYNETTGERGDFPGTYVEYIGRKKISP VVIQD--NSDIKVVPRRKAKIIRD----- Balibase 1 abo Reference 2 1 abo. A -NLFVALYDFVASGDNTLSITKGEKLRVLGYNHN-------GEW 1 ycs. B KGVIYALWDYEPQNDDELPMKEGDCMTIIHREDE DE------IEW 1 pht GYQYRALYDYKKEREEDIDLHLGDILTVNKGSLVALGFSDGQEARPEEIGW 1 ihv. A -NFRVYYRDSRD------PVWKGPAKLLWKG---------EGA * : 1 abo. A 1 ycs. B 1 pht 1 ihv. A CEAQTK--NGQGWVPSNYITPVN-----WWARL--NDKEGYVPRNLLGLYP-----LNGYNe. TTGERGDFPGTYVEYIGRKKISP VVIQD--NSDIKVVPRRKAKIIRD-----

3 D TCoffee. DPA Vs The Human Kinome… l Sequences in our Kinome MSA dataset have been provided by Aventis l Do not inlude the Alpha Kinases l Assembling an exhaustive Kinome Dataset remains a target (c. f. Projects)