Classification trees CART classification regression trees 1 Tree




- Slides: 12

Classification trees CART (classification & regression trees) 1

Tree algorithmbinary recursive partitioning The classification tree or regression tree method examines all values of all the predictors (Xs) and finds the best value of the best predictor that splits the data into two groups (nodes) that are as different as possible on the outcome. This process is independently repeated in the two “daughter” nodes created by the split until either the final (“terminal”) nodes are homogeneous (all observations are the same value, sample size is too small (default: n <5) or the difference between the two nodes is not statistically significant. 2

Example- survival on titanic 3

Survival on the Titanic Outcome (Y): Survival (did not drown) – y/n Predictors (Xs): Gender – M or F Passenger class – 1, 2, 3 Age (years) Can swim - y/n 4

Tree from Titanic data- Y=survival 5

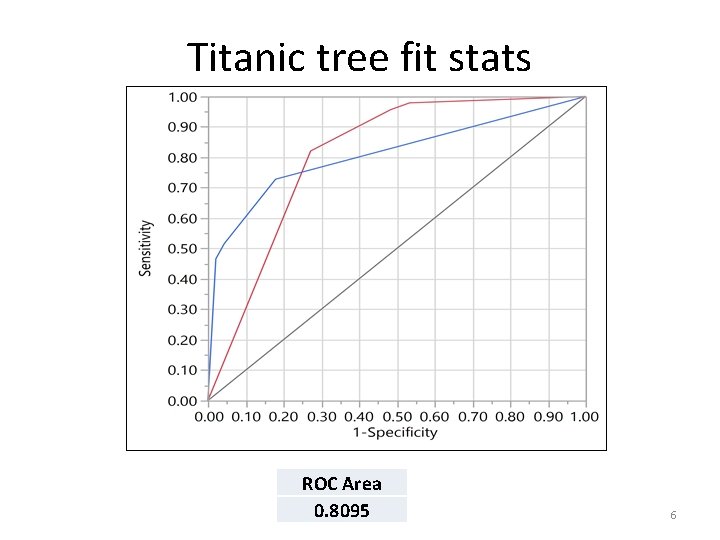
Titanic tree fit stats ROC Area 0. 8095 6

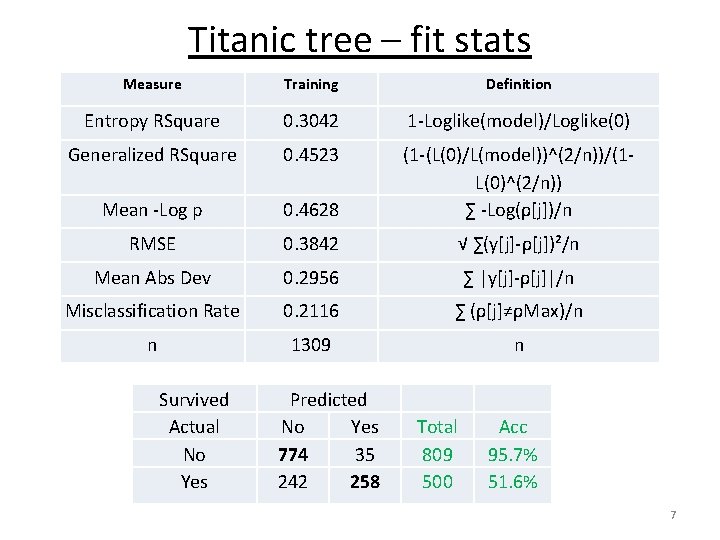
Titanic tree – fit stats Measure Training Definition Entropy RSquare 0. 3042 1 -Loglike(model)/Loglike(0) Generalized RSquare 0. 4523 Mean -Log p 0. 4628 (1 -(L(0)/L(model))^(2/n))/(1 L(0)^(2/n)) ∑ -Log(ρ[j])/n RMSE 0. 3842 √ ∑(y[j]-ρ[j])²/n Mean Abs Dev 0. 2956 ∑ |y[j]-ρ[j]|/n Misclassification Rate 0. 2116 ∑ (ρ[j]≠ρMax)/n n 1309 n Survived Actual No Yes Predicted No Yes 774 35 242 258 Total 809 500 Acc 95. 7% 51. 6% 7

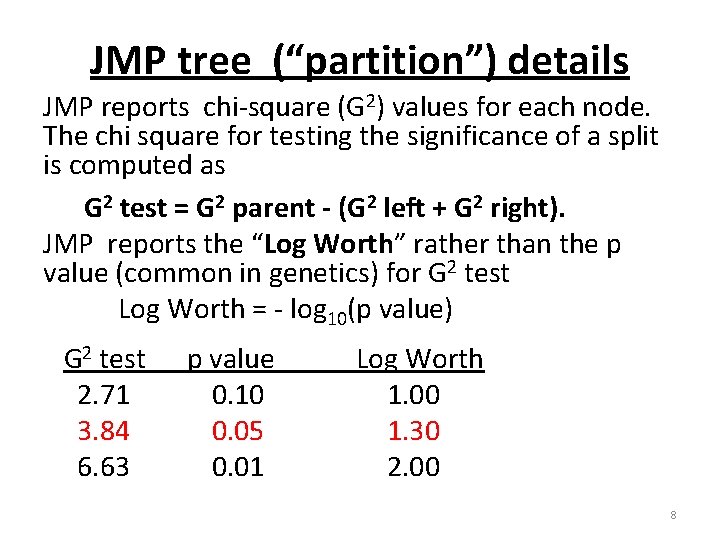
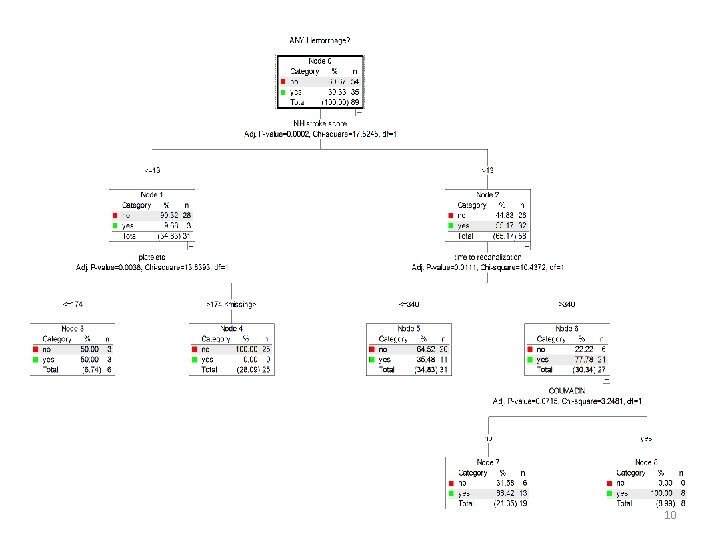
JMP tree (“partition”) details JMP reports chi-square (G 2) values for each node. The chi square for testing the significance of a split is computed as G 2 test = G 2 parent - (G 2 left + G 2 right). JMP reports the “Log Worth” rather than the p value (common in genetics) for G 2 test Log Worth = - log 10(p value) G 2 test 2. 71 3. 84 6. 63 p value 0. 10 0. 05 0. 01 Log Worth 1. 00 1. 30 2. 00 8

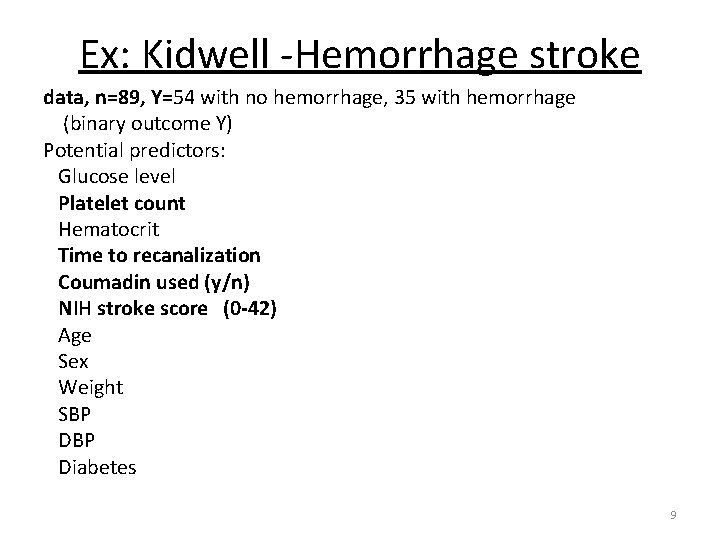
Ex: Kidwell -Hemorrhage stroke data, n=89, Y=54 with no hemorrhage, 35 with hemorrhage (binary outcome Y) Potential predictors: Glucose level Platelet count Hematocrit Time to recanalization Coumadin used (y/n) NIH stroke score (0 -42) Age Sex Weight SBP Diabetes 9

10

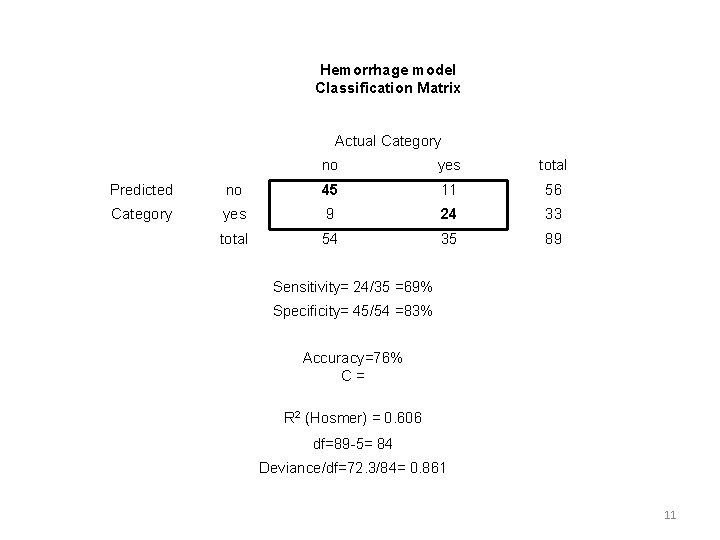
Hemorrhage model Classification Matrix Actual Category no yes total Predicted no 45 11 56 Category yes 9 24 33 total 54 35 89 Sensitivity= 24/35 =69% Specificity= 45/54 =83% Accuracy=76% C= R 2 (Hosmer) = 0. 606 df=89 -5= 84 Deviance/df=72. 3/84= 0. 861 11

JMP partition details (cont) Can automate making the tree with “k fold cross validation” option (k=5 by default). Choose this option, then select “go”. May need to “prune” tree (remove non significant terminal nodes). 12