CLASSIFICATION OF LIFE TAXONOMY Learning Goal I will




- Slides: 13

CLASSIFICATION OF LIFE: TAXONOMY Learning Goal: I will be able to explain the fundamental principles of taxonomy and phylogeny by what a taxon is (list the 7 categories from more general to least specific), who Linnaeus is, define cladistics, phylogeny, clade, node, binomial nomenclature, what the 3 domains are, and the 4 kingdoms within eukarya

Carl Linnaeus developed the scientific naming system still used today. • Taxonomy is the science of naming and classifying organisms. • A taxon is a group of organisms in a classification system. • 7 Taxa (plural for taxon) are: Tyto alba Kingdom Family Phylum Genus Class Species Order Binomial nomenclature (scientific name): is a two-part naming system Genus species (genus always capitalized)

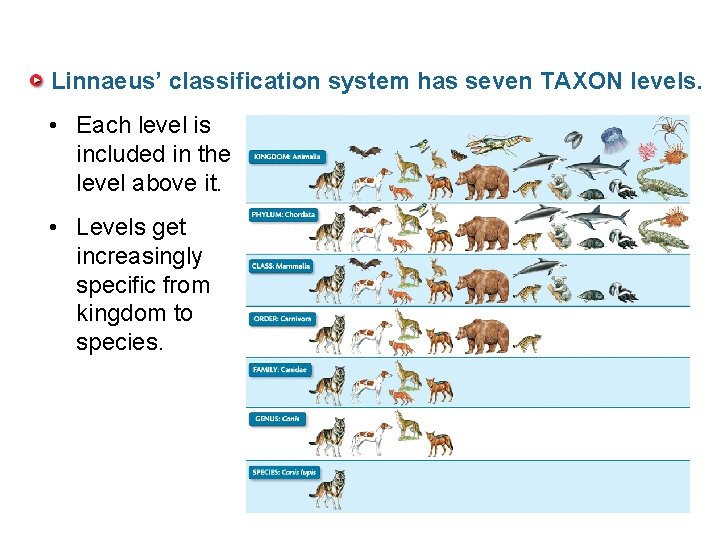
Linnaeus’ classification system has seven TAXON levels. • Each level is included in the level above it. • Levels get increasingly specific from kingdom to species.

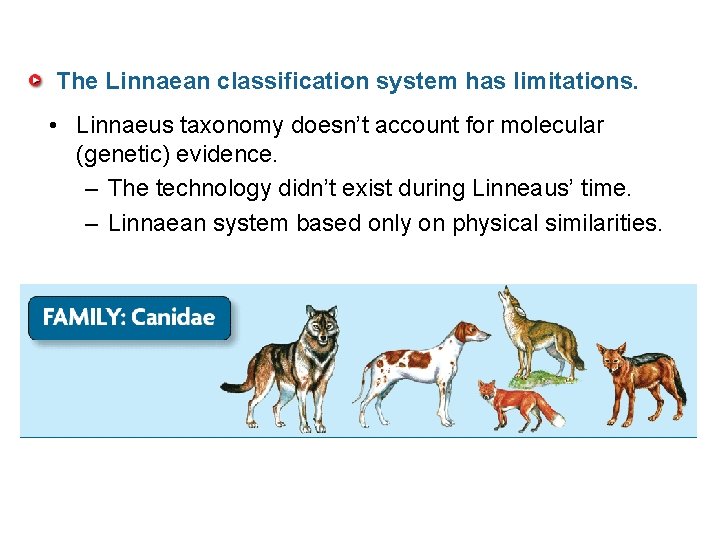
The Linnaean classification system has limitations. • Linnaeus taxonomy doesn’t account for molecular (genetic) evidence. – The technology didn’t exist during Linneaus’ time. – Linnaean system based only on physical similarities.

• Physical similarities are not always the result of close relationships. • Genetic similarities more accurately show evolutionary relationships.

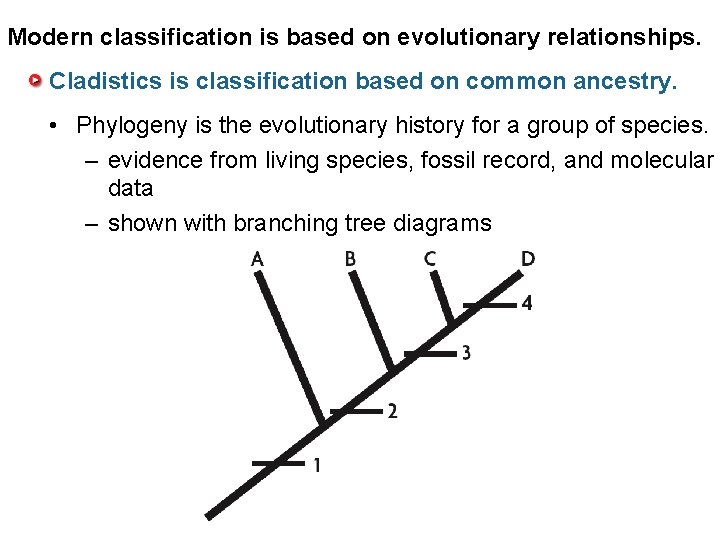
Modern classification is based on evolutionary relationships. Cladistics is classification based on common ancestry. • Phylogeny is the evolutionary history for a group of species. – evidence from living species, fossil record, and molecular data – shown with branching tree diagrams

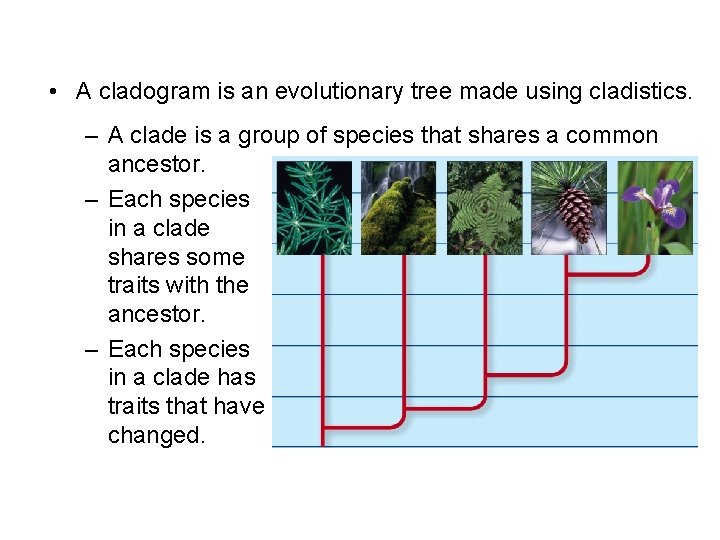
• A cladogram is an evolutionary tree made using cladistics. – A clade is a group of species that shares a common ancestor. – Each species in a clade shares some traits with the ancestor. – Each species in a clade has traits that have changed.

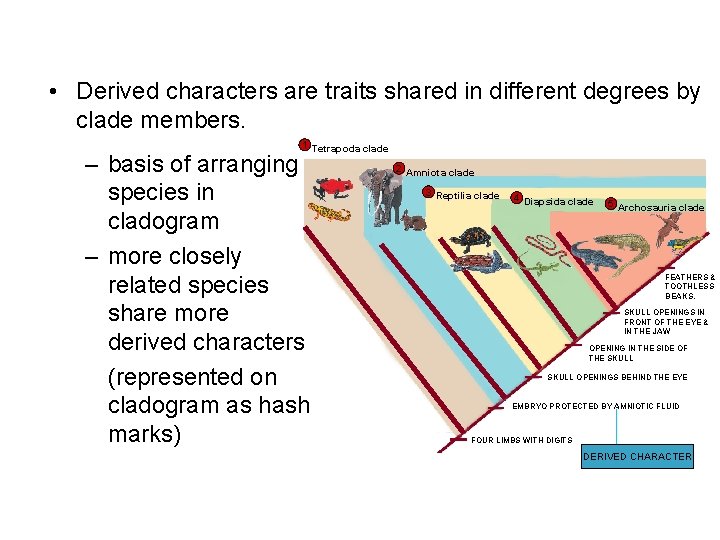
• Derived characters are traits shared in different degrees by clade members. 1 Tetrapoda clade – basis of arranging species in cladogram – more closely related species share more derived characters (represented on cladogram as hash marks) 2 Amniota clade 3 Reptilia clade 4 Diapsida clade 5 Archosauria clade FEATHERS & TOOTHLESS BEAKS. SKULL OPENINGS IN FRONT OF THE EYE & IN THE JAW OPENING IN THE SIDE OF THE SKULL OPENINGS BEHIND THE EYE EMBRYO PROTECTED BY AMNIOTIC FLUID FOUR LIMBS WITH DIGITS DERIVED CHARACTER

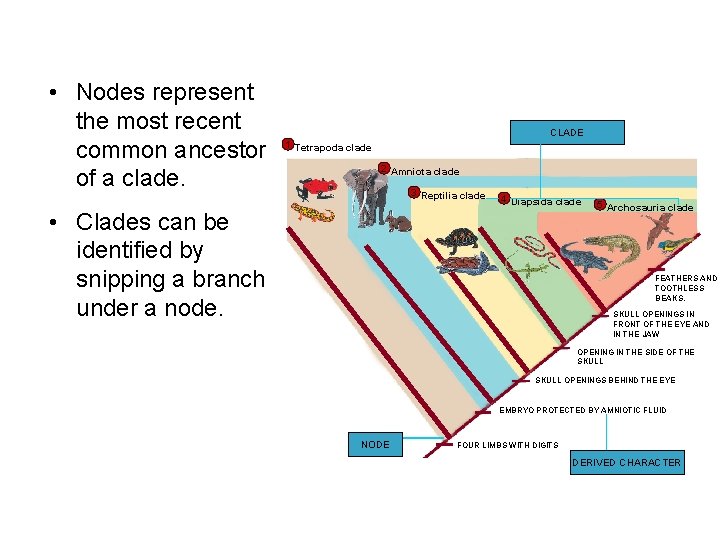
• Nodes represent the most recent common ancestor of a clade. CLADE 1 Tetrapoda clade 2 Amniota clade 3 Reptilia clade • Clades can be identified by snipping a branch under a node. 4 Diapsida clade 5 Archosauria clade FEATHERS AND TOOTHLESS BEAKS. SKULL OPENINGS IN FRONT OF THE EYE AND IN THE JAW OPENING IN THE SIDE OF THE SKULL OPENINGS BEHIND THE EYE EMBRYO PROTECTED BY AMNIOTIC FLUID NODE FOUR LIMBS WITH DIGITS DERIVED CHARACTER

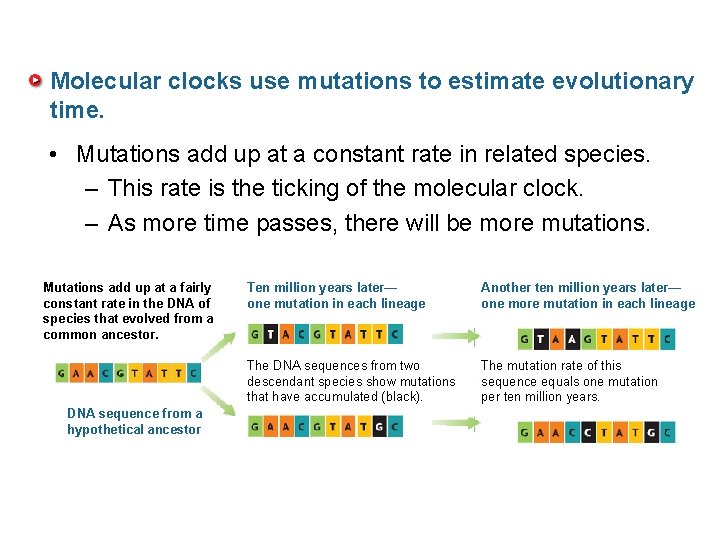
Molecular clocks use mutations to estimate evolutionary time. • Mutations add up at a constant rate in related species. – This rate is the ticking of the molecular clock. – As more time passes, there will be more mutations. Mutations add up at a fairly constant rate in the DNA of species that evolved from a common ancestor. DNA sequence from a hypothetical ancestor Ten million years later— one mutation in each lineage Another ten million years later— one more mutation in each lineage The DNA sequences from two descendant species show mutations that have accumulated (black). The mutation rate of this sequence equals one mutation per ten million years.

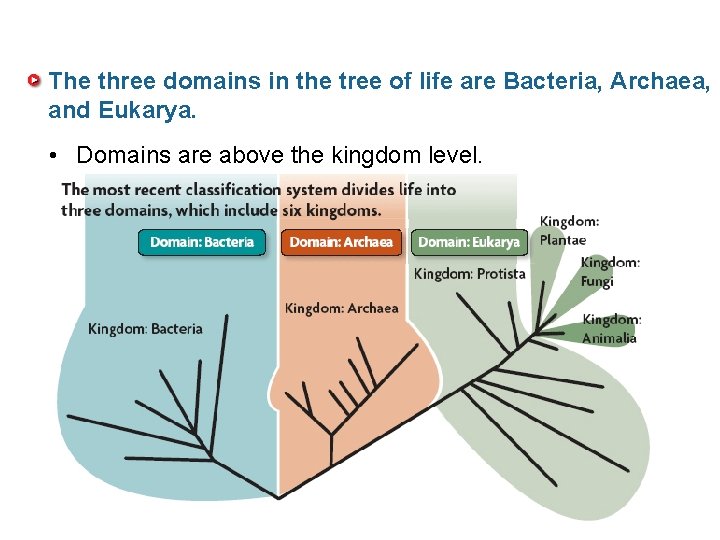
The three domains in the tree of life are Bacteria, Archaea, and Eukarya. • Domains are above the kingdom level.

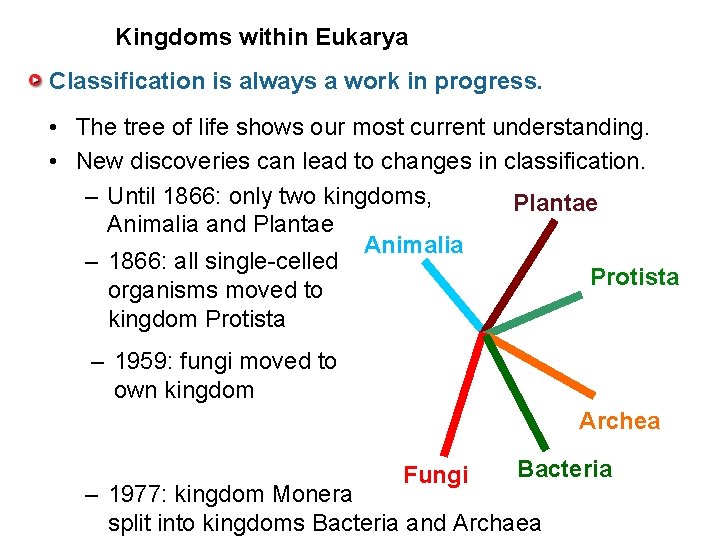
Kingdoms within Eukarya Classification is always a work in progress. • The tree of life shows our most current understanding. • New discoveries can lead to changes in classification. – Until 1866: only two kingdoms, Plantae Animalia and Plantae Animalia – 1866: all single-celled Protista organisms moved to kingdom Protista – 1959: fungi moved to own kingdom Archea Fungi Bacteria – 1977: kingdom Monera split into kingdoms Bacteria and Archaea

HOW DID WE DO Learning Goal: I will be able to explain the fundamental principles of taxonomy and phylogeny by what a taxon is (list the 7 categories from more general to least specific), who Linnaeus is, define cladistics, phylogeny, clade, node, binomial nomenclature, what the 3 domains are, and the 4 kingdoms within eukarya