Classification Analysis of HIV RNase H Bioassay Lianyi

Classification Analysis of HIV RNase H Bioassay Lianyi Han Computational Biology Branch NCBI/NLM/NIH Rocky ‘ 07 December 1, 2007 1

Introduction q The need for new anti-HIV agents q q Drug resistant mutations Side effect / Toxicity The limit in virtual screening techniques q Huge chemical space q Structure and activities The challenge to generate new hypothesis q q Noise reduction Knowledge exploration December, 2007 2
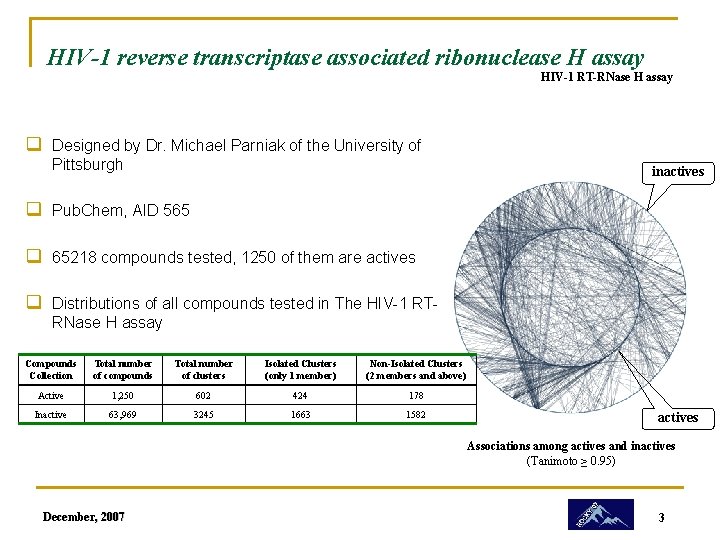
HIV-1 reverse transcriptase associated ribonuclease H assay HIV-1 RT-RNase H assay q Designed by Dr. Michael Parniak of the University of Pittsburgh inactives q Pub. Chem, AID 565 q 65218 compounds tested, 1250 of them are actives q Distributions of all compounds tested in The HIV-1 RTRNase H assay Compounds Collection Total number of compounds Total number of clusters Isolated Clusters (only 1 member) Non-Isolated Clusters (2 members and above) Active 1, 250 602 424 178 Inactive 63, 969 3245 1663 1582 actives Associations among actives and inactives (Tanimoto ≥ 0. 95) December, 2007 3
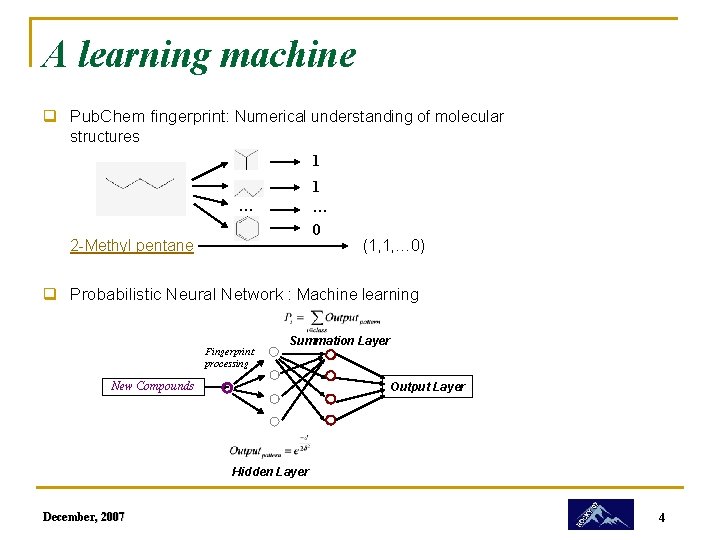
A learning machine q Pub. Chem fingerprint: Numerical understanding of molecular structures 1 1 … 0 … 2 -Methyl pentane (1, 1, … 0) q Probabilistic Neural Network : Machine learning Fingerprint processing Summation Layer New Compounds Output Layer Hidden Layer December, 2007 4
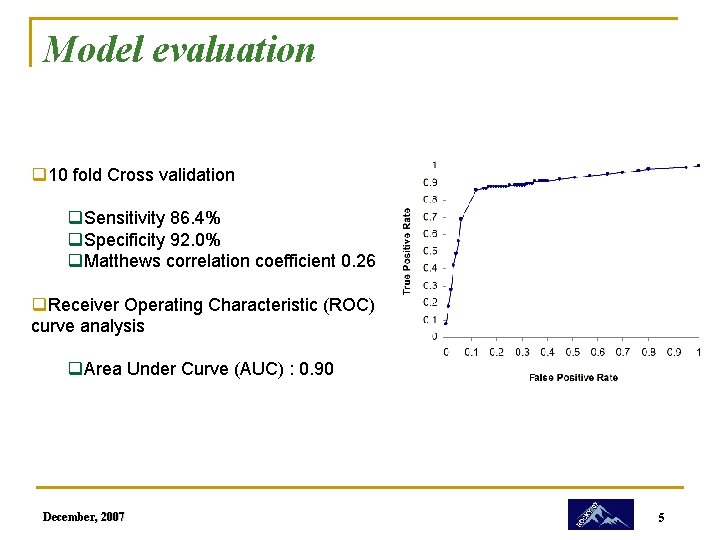
Model evaluation q 10 fold Cross validation q. Sensitivity 86. 4% q. Specificity 92. 0% q. Matthews correlation coefficient 0. 26 q. Receiver Operating Characteristic (ROC) curve analysis q. Area Under Curve (AUC) : 0. 90 December, 2007 5
Conclusions q The bioactivity data of HIV-1 RT-RNH assay can be learned for new hypothesis q The machine learning of HTS data can be used for virtual hits exploration Acknowledgements q Yanli Wang q Steve Bryant q This research was supported by the Intramural Research Program of the NIH/NLM December, 2007 6
- Slides: 6