Classic paper Goal was to automate DNA sequencing


- Slides: 8

• Classic paper – Goal was to automate DNA sequencing to facilitate sequencing of human genome – Describes new form of sequencing using two-step PCR to generate long, even reads • First step amplifies template DNA by PCR • Second step does asymmetric amplification (linear) to perform actual sequencing reaction • Proof-of-principle for automated sequencing Bio. Sci 145 B week 4 papers page 1 ©copyright Bruce Blumberg 2004. All rights reserved

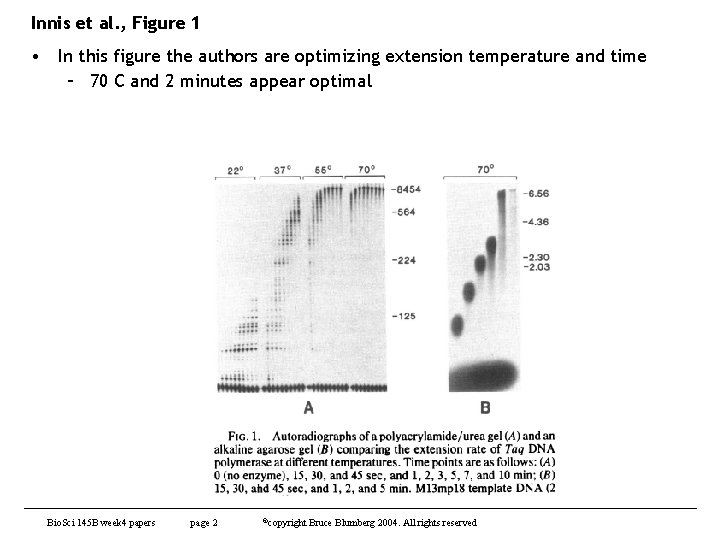
Innis et al. , Figure 1 • In this figure the authors are optimizing extension temperature and time – 70 C and 2 minutes appear optimal Bio. Sci 145 B week 4 papers page 2 ©copyright Bruce Blumberg 2004. All rights reserved

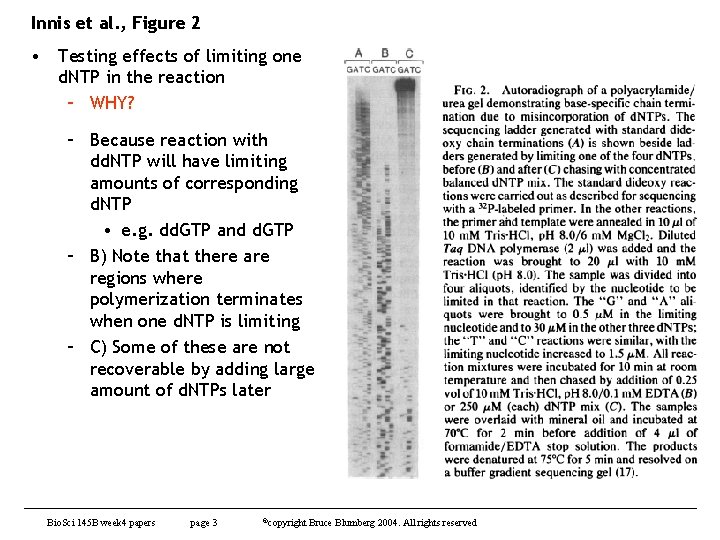
Innis et al. , Figure 2 • Testing effects of limiting one d. NTP in the reaction – WHY? – Because reaction with dd. NTP will have limiting amounts of corresponding d. NTP • e. g. dd. GTP and d. GTP – B) Note that there are regions where polymerization terminates when one d. NTP is limiting – C) Some of these are not recoverable by adding large amount of d. NTPs later Bio. Sci 145 B week 4 papers page 3 ©copyright Bruce Blumberg 2004. All rights reserved

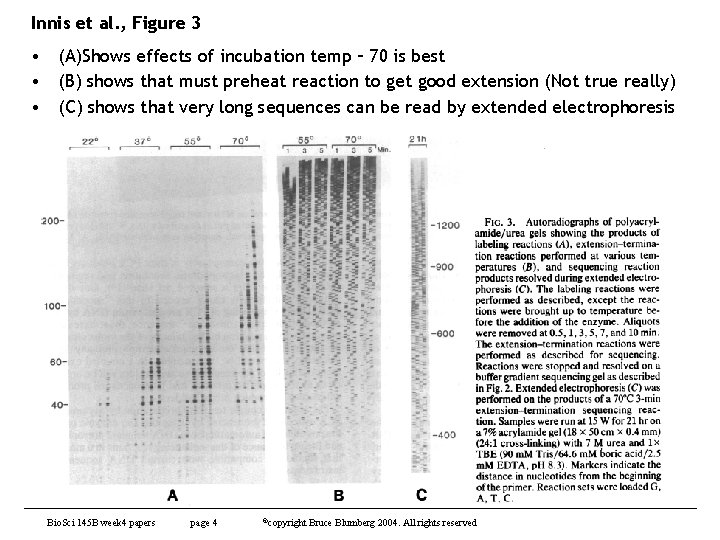
Innis et al. , Figure 3 • (A)Shows effects of incubation temp – 70 is best • (B) shows that must preheat reaction to get good extension (Not true really) • (C) shows that very long sequences can be read by extended electrophoresis Bio. Sci 145 B week 4 papers page 4 ©copyright Bruce Blumberg 2004. All rights reserved

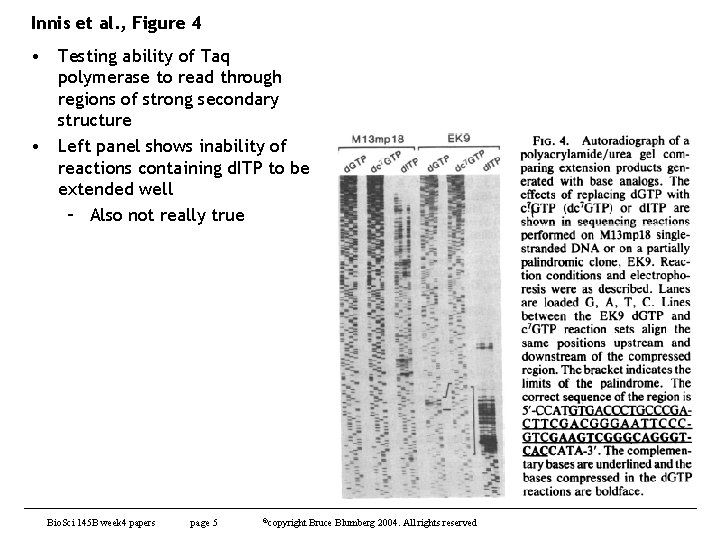
Innis et al. , Figure 4 • Testing ability of Taq polymerase to read through regions of strong secondary structure • Left panel shows inability of reactions containing d. ITP to be extended well – Also not really true Bio. Sci 145 B week 4 papers page 5 ©copyright Bruce Blumberg 2004. All rights reserved

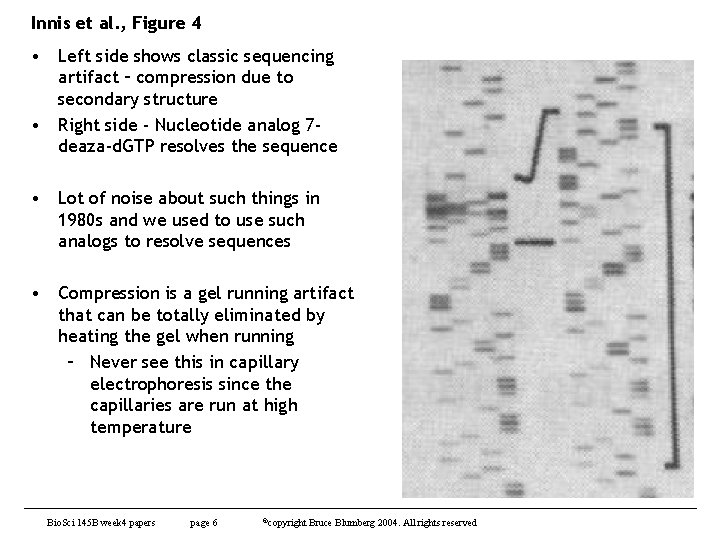
Innis et al. , Figure 4 • Left side shows classic sequencing artifact – compression due to secondary structure • Right side - Nucleotide analog 7 deaza-d. GTP resolves the sequence • Lot of noise about such things in 1980 s and we used to use such analogs to resolve sequences • Compression is a gel running artifact that can be totally eliminated by heating the gel when running – Never see this in capillary electrophoresis since the capillaries are run at high temperature Bio. Sci 145 B week 4 papers page 6 ©copyright Bruce Blumberg 2004. All rights reserved

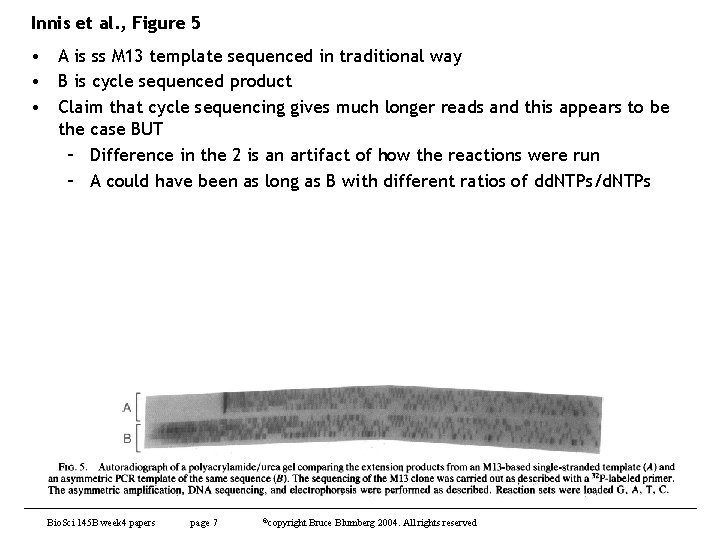
Innis et al. , Figure 5 • A is ss M 13 template sequenced in traditional way • B is cycle sequenced product • Claim that cycle sequencing gives much longer reads and this appears to be the case BUT – Difference in the 2 is an artifact of how the reactions were run – A could have been as long as B with different ratios of dd. NTPs/d. NTPs Bio. Sci 145 B week 4 papers page 7 ©copyright Bruce Blumberg 2004. All rights reserved

Innis et al. • Conclusions drawn – Taq polymerase is good for sequencing • Long reads (processive) • Even banding patterns (no 3’ exonuclease activity) • Incorporates nucleotide analogs to improve sequencing gels – Developed new sequencing method • Optimized temperature and time for • Very suitable for automated sequencing • Value of the method – This is how all sequencing is done today (with labeled dd. NTPs) – Advances in template prep (Templi. Phi) and – Terminator chemistry (Big Dyes) – Have led to rapid completion of genome projects ahead of schedule and under budget Bio. Sci 145 B week 4 papers page 8 ©copyright Bruce Blumberg 2004. All rights reserved