Class 4 Fast Sequence Alignment Alignment in Real



![Banded DP that we have two strings s[1. . n] and t[1. . m] Banded DP that we have two strings s[1. . n] and t[1. . m]](https://slidetodoc.com/presentation_image_h/f4b4e1cb59da8d2700764d8a3f765b88/image-5.jpg)
![Banded DP Problem (for local alignment): u If we know that t[i. . j] Banded DP Problem (for local alignment): u If we know that t[i. . j]](https://slidetodoc.com/presentation_image_h/f4b4e1cb59da8d2700764d8a3f765b88/image-7.jpg)



- Slides: 14

Class 4: Fast Sequence Alignment .

Alignment in Real Life u One of the major uses of alignments is to find sequences in a “database” u Such collections contain massive number of sequences (order of 106) u Finding homologies in these databases with the standard dynamic programming can take too long

Heuristic Search u Instead, most searches rely on heuristic procedures u These are not guaranteed to find the best match u Sometimes, they will completely miss a highscoring match u We now describe the main ideas used by some of these procedures · Actual implementations often contain additional tricks and hacks

Basic Intuition u The main resource consuming factor in the standard DP is decision of where the gaps are. If there were no gaps, life was easy! u Almost all heuristic search procedures are based on the observation that real-life well-matching pairs of sequences often do contain long strings with gapless matches. u These heuristics try to find significant local gap-less matches and then extend them.
![Banded DP that we have two strings s1 n and t1 m Banded DP that we have two strings s[1. . n] and t[1. . m]](https://slidetodoc.com/presentation_image_h/f4b4e1cb59da8d2700764d8a3f765b88/image-5.jpg)
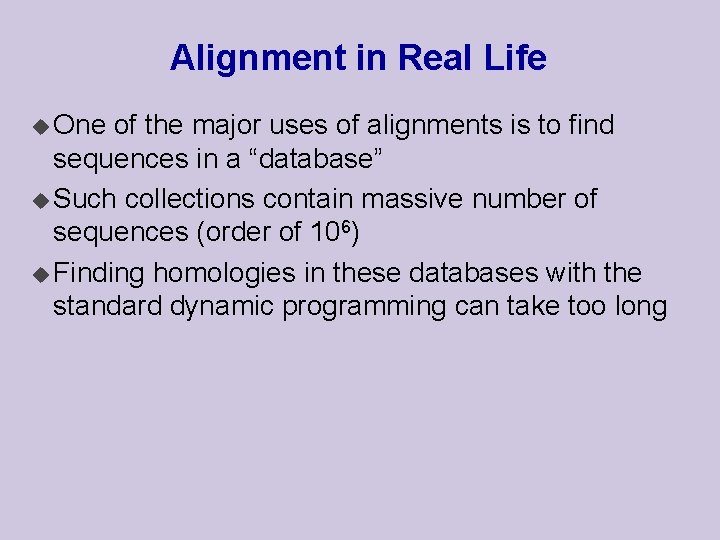
Banded DP that we have two strings s[1. . n] and t[1. . m] such that n m u If the optimal global alignment of s and t has few gaps, then path of the alignment will be close to the diagonal s u Suppose t

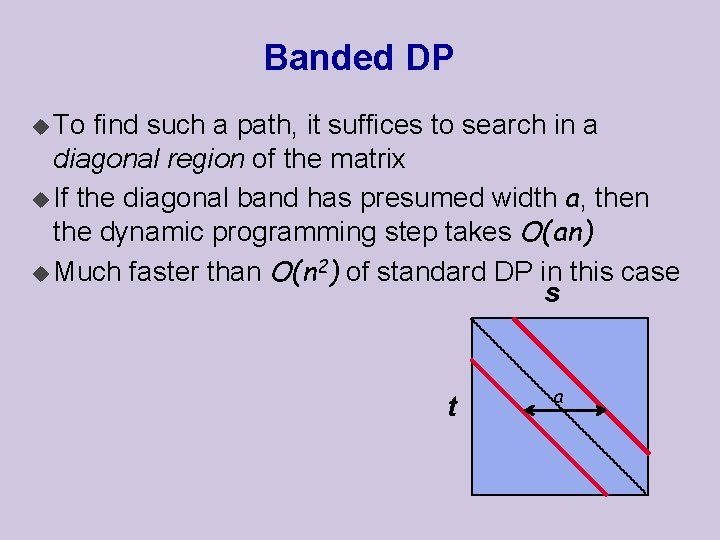
Banded DP u To find such a path, it suffices to search in a diagonal region of the matrix u If the diagonal band has presumed width a, then the dynamic programming step takes O(an) u Much faster than O(n 2) of standard DP in this case s t a
![Banded DP Problem for local alignment u If we know that ti j Banded DP Problem (for local alignment): u If we know that t[i. . j]](https://slidetodoc.com/presentation_image_h/f4b4e1cb59da8d2700764d8a3f765b88/image-7.jpg)
Banded DP Problem (for local alignment): u If we know that t[i. . j] matches the query s[p. . q], then we can use banded DP to evaluate quality of the match u However, we do not know i, j, p, q ! u How do we select which sub-sequences to align using banded DP?

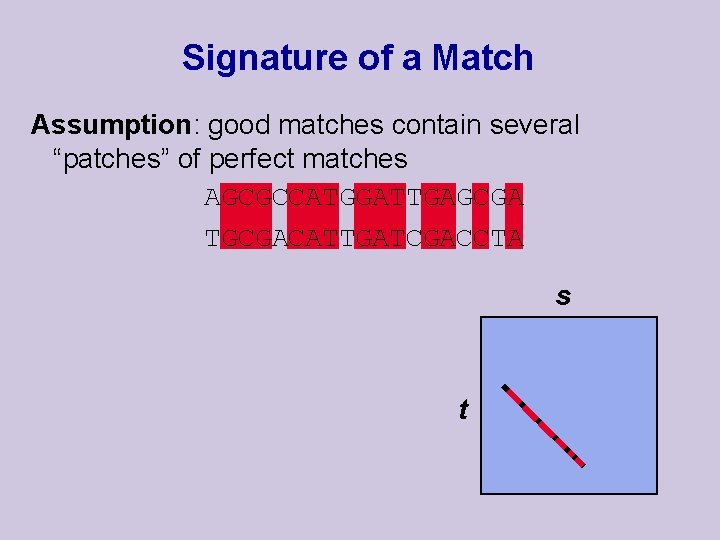
FASTA Overview u Main idea: Find (fast!) “good” diagonals and extend them to complete matches u Suppose that we have a relatively long gap-less local match (diagonal): …AGCGCCATGGATTGAGCGA… …TGCGACATTGATCGACCTA… u Can we find “clues” that will let us find it quickly?

Signature of a Match Assumption: good matches contain several “patches” of perfect matches AGCGCCATGGATTGAGCGA TGCGACATTGATCGACCTA s t

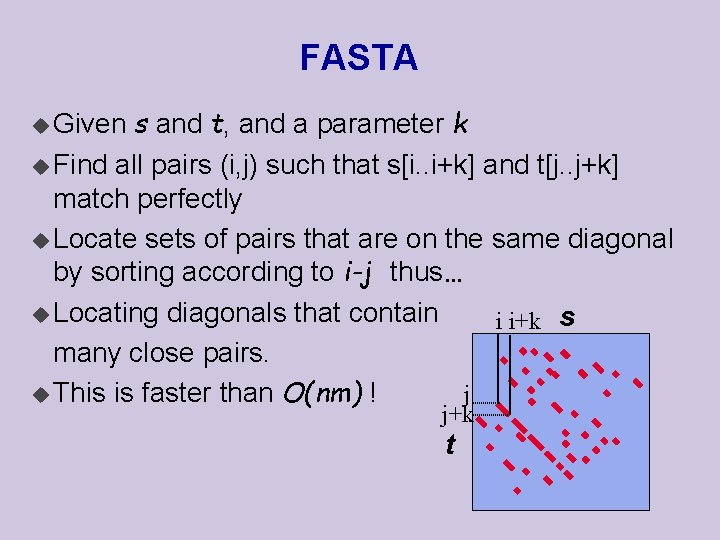
FASTA s and t, and a parameter k u Find all pairs (i, j) such that s[i. . i+k] and t[j. . j+k] match perfectly u Locate sets of pairs that are on the same diagonal by sorting according to i-j thus… u Locating diagonals that contain i i+k s many close pairs. j u This is faster than O(nm) ! u Given j+k t

FASTA u Extend the “best” diagonal matches to imperfect (yet ungapped) matches, compute alignment scores per diagonal. Pick the best-scoring matches. u Try to combine close diagonals to potential gapped matches, picking the best-scoring matches. u Finally, run banded DP on the regions containing these matches, resulting in several good candidate alignments. applications of FASTA use very small k (2 for proteins, and 4 -6 for DNA) u Most

BLAST Overview u FASTA drawback is its reliance on perfect matches u BLAST (Basic Local Alignment Search Tool)uses similar intuition, but relies on high scoring matches rather than exact matches parameters: length k, and threshold T u Two strings s and t of length k are a high scoring pair (HSP) if d(s, t) > T u Given

High-Scoring Pair a query string s, BLAST construct all words w (“neighborhood words”), such that w is an HSP with a k-substring of s. · Note that not all k-mers have an HSP in s u Search the database for perfect matches with neighborhood words. Those are “hits” for further alignment. · We can locate seed words in a large database in a single pass, given the database is properly preprocessed (using hashing techniques). u Given

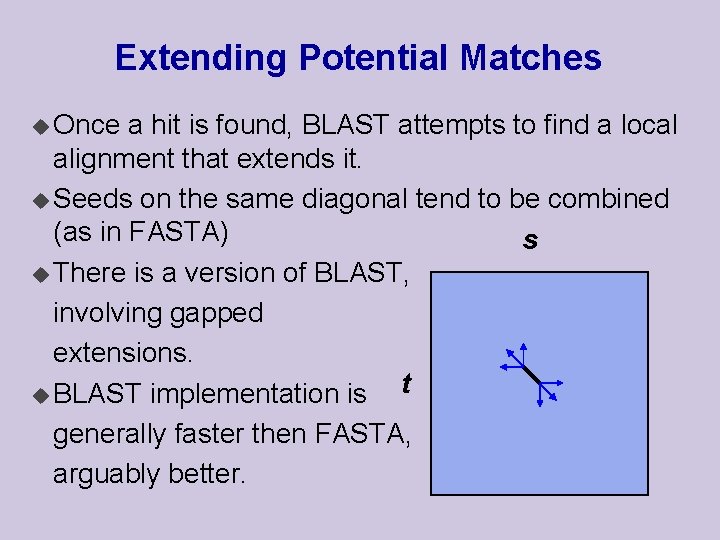
Extending Potential Matches u Once a hit is found, BLAST attempts to find a local alignment that extends it. u Seeds on the same diagonal tend to be combined (as in FASTA) s u There is a version of BLAST, involving gapped extensions. t u BLAST implementation is generally faster then FASTA, arguably better.