Clasificacin molecular del colangiocarcinoma extraheptico Robert Montal Wei

Clasificación molecular del colangiocarcinoma extrahepático Robert Montal, Wei Qiang Leow, Carla Montironi, Laia Bassaganyas, Agrin Moeini, Daniela Sia, Roser Pinyol, Judit Peix, Augusto Villanueva, Josep M Llovet. #SEOM 2018

Disclosure Information q Employment: IDIBAPS, Fundació Clínic. q Consultant or Advisory Role: q Stock Ownership: q Research Funding: European Commission/Horizon 2020 Program (HEPCAR, Ref 667273 -2), Asociación Española Contra el Cáncer (AECC), Spanish National Health Institute (SAF 2016 -76390) and the Generalitat de Catalunya/AGAUR (SGR-1162 and SGR-1358). q Speaking: q Grant support: SEOM. q Other: #SEOM 2018
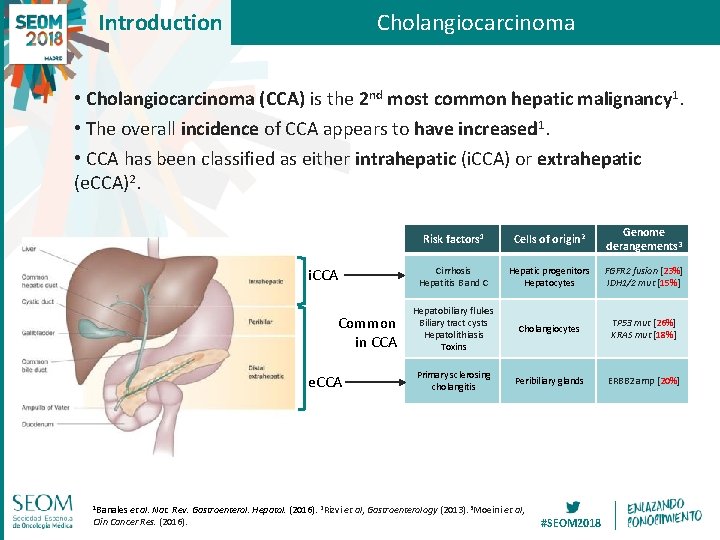
Introduction Cholangiocarcinoma • Cholangiocarcinoma (CCA) is the 2 nd most common hepatic malignancy 1. • The overall incidence of CCA appears to have increased 1. • CCA has been classified as either intrahepatic (i. CCA) or extrahepatic (e. CCA)2. i. CCA Common in CCA e. CCA 1 Banales et Risk factors 1 Cells of origin 2 Genome derangements 3 Cirrhosis Hepatitis B and C Hepatic progenitors Hepatocytes FGFR 2 fusion [23%] IDH 1/2 mut [15%] Hepatobiliary flukes Biliary tract cysts Hepatolithiasis Toxins Cholangiocytes TP 53 mut [26%] KRAS mut [18%] Primary sclerosing cholangitis Peribiliary glands ERBB 2 amp [20%] al. Nat. Rev. Gastroenterol. Hepatol. (2016). 2 Rizvi et al, Gastroenterology (2013). 3 Moeini et al, Clin Cancer Res. (2016). #SEOM 2018
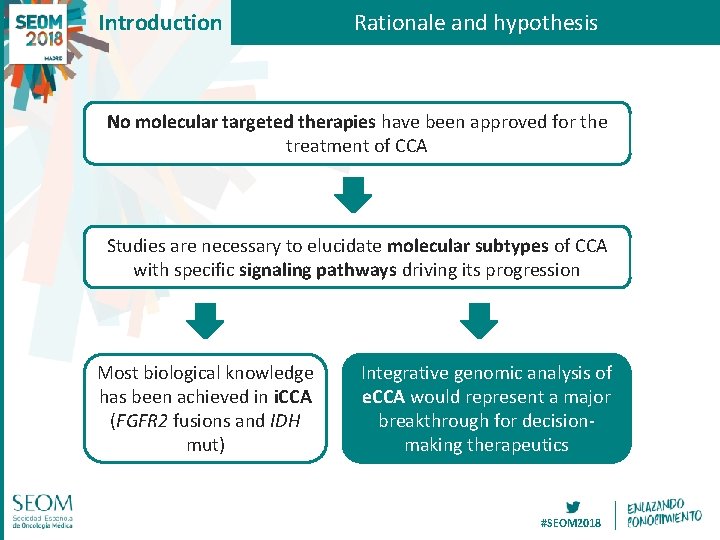
Introduction Rationale and hypothesis No molecular targeted therapies have been approved for the treatment of CCA Studies are necessary to elucidate molecular subtypes of CCA with specific signaling pathways driving its progression Most biological knowledge has been achieved in i. CCA (FGFR 2 fusions and IDH mut) Integrative genomic analysis of e. CCA would represent a major breakthrough for decisionmaking therapeutics #SEOM 2018
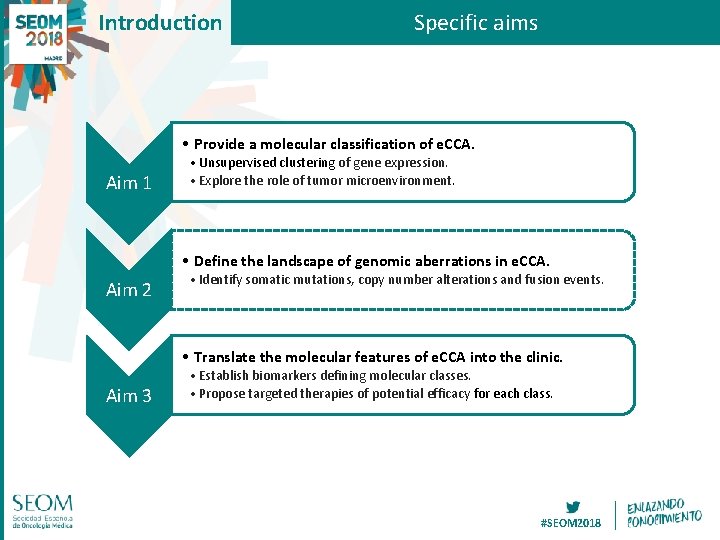
Introduction Specific aims • Provide a molecular classification of e. CCA. Aim 1 • Unsupervised clustering of gene expression. • Explore the role of tumor microenvironment. • Define the landscape of genomic aberrations in e. CCA. Aim 2 • Identify somatic mutations, copy number alterations and fusion events. • Translate the molecular features of e. CCA into the clinic. Aim 3 • Establish biomarkers defining molecular classes. • Propose targeted therapies of potential efficacy for each class. #SEOM 2018

Methods Flow chart of the study Patients recruited (N=208) Collaborating center Mount Sinai (New York) [N=59] Clínic (Barcelona) [N=39] CHUV (Lausanne) [N=36] Mayo Clinic (Rochester) [N=28] Lenox Hill (New York) [N=19] Johns Hokpkins (Baltimore) [N=15] Vall Hebron (Barcelona) [N=12] Surgically resected e. CCA Patients excluded (N=19) (14 i. CCA, 4 non-resected e. CCA, 1 non-viable e. CCA) Patients included (N=189) Tumor macrodissection RNA Whole genome expression (N=182) Unsupervised clustering DNA Targeted exome sequencing (N=163) Unstained slides Immunohistochemistry (ERBB 2, PD 1, PD-L 1) (N=182) Clinicalpathological characteristics Integrative characterization #SEOM 2018

Results Transcriptome-based unsupervised clustering Normalization (RMA) Batch correction (Combat) Metabolic (18. 7%) Remove noise (Preprocess) Proliferation (22. 5%) Clustering (NMF consensus) Mesenchymal (47. 3%) Immune (11. 5%) #SEOM 2018

Results Biological hallmarks / Tumor microenvironment p=0. 002 p<0. 001 #SEOM 2018
Results Clinical/pathological/molecular associations #SEOM 2018

Results Metabolic class (18. 7%) Cluster 1 CCA Andersen (p<0. 001) S 3 class HCC Hoshida (p<0. 001) CTNNB 1 class HCC Chiang (p<0. 001) Absence of Immune class HCC Sia (p=0. 038) Absence of Active stroma class PDAC Moffitt (p<0. 001) #SEOM 2018

Results Proliferation class (22. 5%) Papillary histology (p=0. 004) Intraductal papillary neoplasm of the bile duct (p=0. 017) Higher tumor purity (p=0. 001) Proliferation class i. CCA Sia (p<0. 001) S 2 class HCC Hoshida (p<0. 001) Proliferation class HCC Chiang (p<0. 001) Classical class PDAC Moffiftt (p<0. 001) Classic class PDAC Collisson (p=0. 004) #SEOM 2018
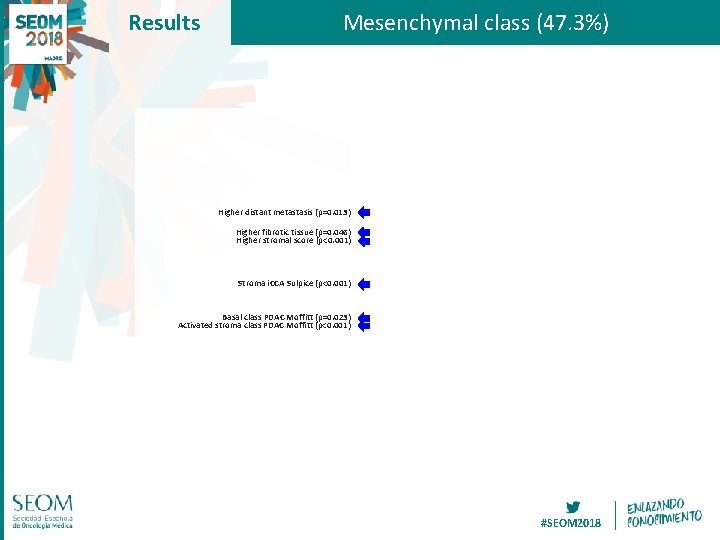
Results Mesenchymal class (47. 3%) Higher distant metastasis (p=0. 013) Higher fibrotic tissue (p=0. 046) Higher stromal score (p<0. 001) Stroma i. CCA Sulpice (p<0. 001) Basal class PDAC Moffitt (p=0. 023) Activated stroma class PDAC Moffitt (p<0. 001) #SEOM 2018

Results Immune class (11. 5%) Higher lymphocyte infiltration (p=0. 001) Higher immune score (p=0. 002) Immune class HCC Sia (p=0. 017) #SEOM 2018
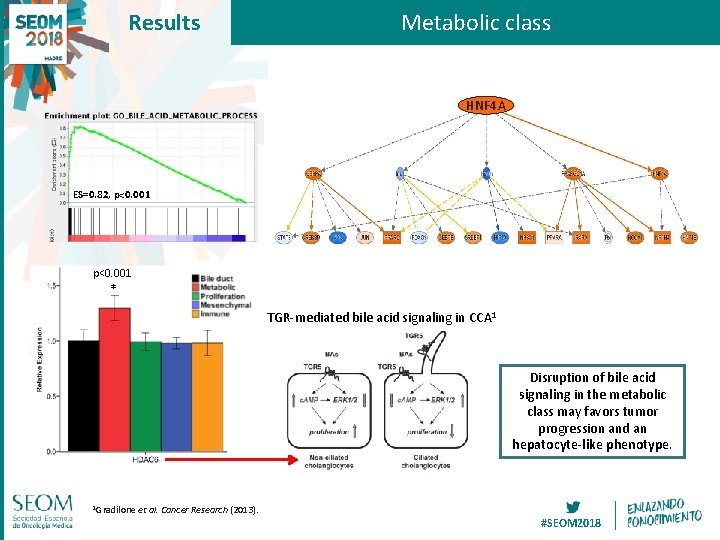
Results Metabolic class HNF 4 A ES=0. 82, p<0. 001 * TGR-mediated bile acid signaling in CCA 1 Disruption of bile acid signaling in the metabolic class may favors tumor progression and an hepatocyte-like phenotype. 1 Gradilone et al. Cancer Research (2013). #SEOM 2018

Results Proliferation class i. CCA 1 ERBB 2 Subclass mapping Proliferation=31% Rest=9% p=0. 024 Mesenchymal Metabolic e. CCA Immune p=0. 008 Proliferation Inflammation Proliferation ES=0. 60, p=0. 028 1 Sia et ES=0. 59, p=0. 011 al. Gastroenterology (2013). ES=0. 52, p=0. 014 The transcriptome of the e. CCA proliferation class is similar to the proliferation subclass described previously in i. CCA 1, with activation of oncogenic RTK such as ERBB 2. #SEOM 2018
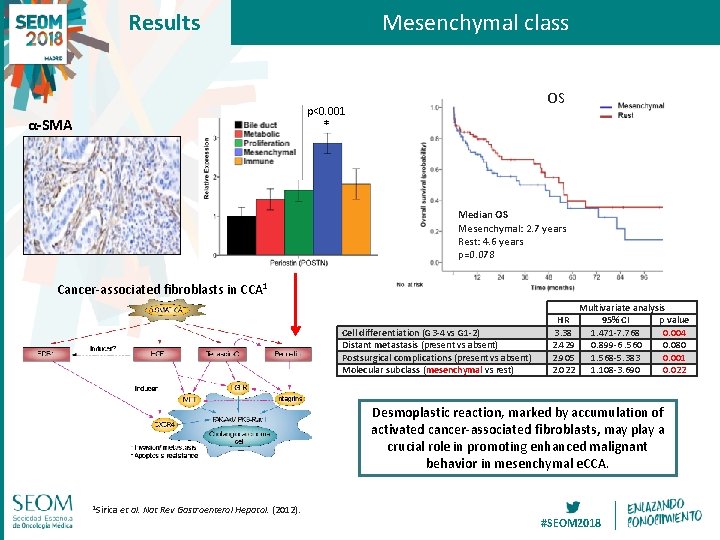
Results Mesenchymal class OS p<0. 001 α-SMA * Median OS Mesenchymal: 2. 7 years Rest: 4. 6 years p=0. 078 Cancer-associated fibroblasts in CCA 1 Cell differentiation (G 3 -4 vs G 1 -2) Distant metastasis (present vs absent) Postsurgical complications (present vs absent) Molecular subclass (mesenchymal vs rest) HR 3. 38 2. 429 2. 905 2. 022 Multivariate analysis 95% CI p value 1. 471 -7. 768 0. 004 0. 899 -6. 560 0. 080 1. 568 -5. 383 0. 001 1. 108 -3. 690 0. 022 Desmoplastic reaction, marked by accumulation of activated cancer-associated fibroblasts, may play a crucial role in promoting enhanced malignant behavior in mesenchymal e. CCA. 1 Sirica et al. Nat Rev Gastroenterol Hepatol. (2012). #SEOM 2018

Results Immune class PD 1 treatment in solid tumors (N=65)1 Subclass mapping Mesenchymal Metabolic Proliferation Immune p=0. 024 Objective No response PD 1 Immune=80% Rest=39% p=0. 001 1 Prat et PDL 1 al. Cancer Research. (2017). Immune=62% Rest=29% p=0. 005 The analysis of immune subpopulations by e. CCA class confirms a higher presence of CD 8 T cells in the immune class, with higher expression of PD 1 PDL 1 immune checkpoints. #SEOM 2018
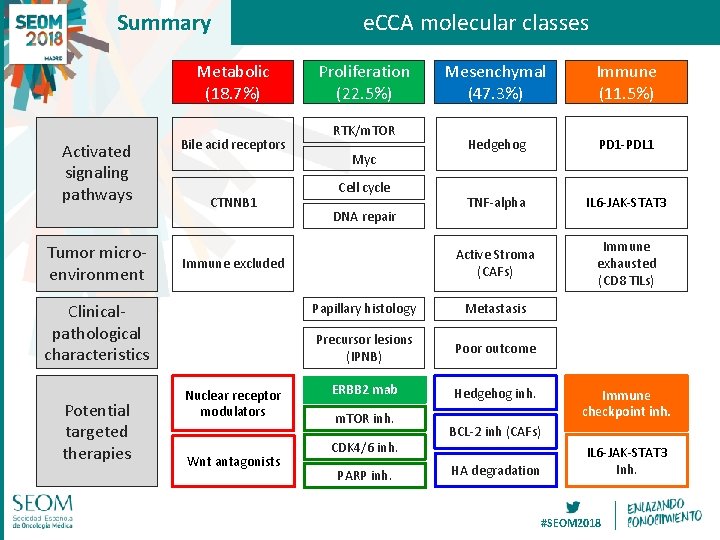
Summary Metabolic (18. 7%) Activated signaling pathways Tumor microenvironment Bile acid receptors CTNNB 1 Proliferation (22. 5%) RTK/m. TOR Myc Cell cycle DNA repair Immune excluded Clinicalpathological characteristics Potential targeted therapies e. CCA molecular classes Nuclear receptor modulators Wnt antagonists Mesenchymal (47. 3%) Immune (11. 5%) Hedgehog PD 1 -PDL 1 TNF-alpha IL 6 -JAK-STAT 3 Active Stroma (CAFs) Immune exhausted (CD 8 TILs) Papillary histology Metastasis Precursor lesions (IPNB) Poor outcome ERBB 2 mab Hedgehog inh. m. TOR inh. CDK 4/6 inh. PARP inh. Immune checkpoint inh. BCL-2 inh (CAFs) HA degradation IL 6 -JAK-STAT 3 Inh. #SEOM 2018
Conclusions Transcriptome-based subtyping of e. CCA identifies four distinct molecular classes that correlate with clinical-pathological characteristics, enhancing the opportunities for therapeutic development. Tumors classified within the metabolic class are enriched by gene signatures defining bile acid metabolism and present overexpression of classic hepatocyte markers. The proliferation class is associated with papillary histology and presents enrichment of MYC, m. TOR and ERBB 2 signaling. The mesenchymal class is associated with signatures defining epithelial-mesenchymal transition as well as stromal activation, with the poorest clinical outcome. Tumors classified as immune class present increased tumor-infiltrating lymphocytes and are characterized by enrichment of PD-1/PD-L 1 signaling. #SEOM 2018

Acknowledgements Liver Cancer Translational Research Laboratory Grant support: Collaborating centers: #SEOM 2018
- Slides: 20