CIRCADIAN RHYTHMS IN PLANTS WHAT IS A CIRCADIAN













- Slides: 28

CIRCADIAN RHYTHMS IN PLANTS * WHAT IS A CIRCADIAN CLOCK? * CLOCK OUTPUTS * CLOCK INPUTS * THE CENTRAL OSCILLATOR * OTHER COMPONENTS * SEASONAL RHYTHMS * EVOLUTIONARY RELATIONSHIPS

WHAT IS A CIRCADIAN CLOCK? * Rhythmicity in behavior, physiology and biochemistry of organisms * Some rhythms persist even without environmental changes Ø circadian rhythms – 24 h period Ø endogenous control: circadian clock * Anticipation of rhythmic changes in the environment Ø changes in the physiological state that provide them with an adaptive advantage RHYTHMIC OUTPUTS - PROCESSES REGULATED BY THE CLOCK * de Mairan, 1727: rhythmic leaf movements in constant darkness * most of current knowledge based on rhythmic cellular or physiological processes: a) mammals: digestion, regulation of body temperature, hormone secretion, time of sleep onset b) cyanobacteria: photosynthesis, nitrogen fixation, cell division

RHYTHMIC OUTPUTS c) plants: leaf movements, cell elongation rates, stomatal aperture, CO 2 assimilation, Calvin cycle, ethylene production, hypocotyl elongation… * Gene expression: a) cyanobacteria: 80% of genes are CCGs > the clock regulates the transcriptional machinery? > additional regulatory layers for many genes b) Arabidopsis: 6% are CCGs, peaking at all phases of day and night CCG clusters * photosynthesis-related genes * photoreceptor genes and downstream signaling components * photoprotective pigments * chilling resistance, cold and drought * carbon allocation, nitrogen and sulfur assimilation * flowering, cell elongation * 25% of the CCGs are totally uncharacterized!!! very conserved motif, “evening element”, present 46 times in the promoters of 31 genes

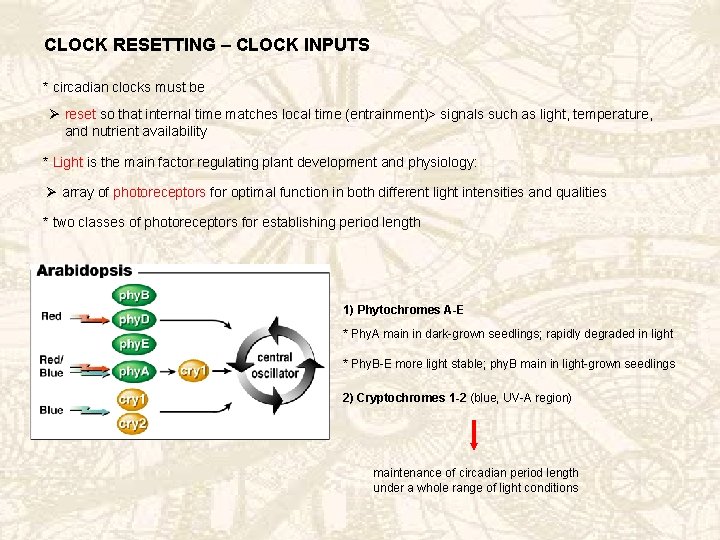
CLOCK RESETTING – CLOCK INPUTS * circadian clocks must be Ø reset so that internal time matches local time (entrainment)> signals such as light, temperature, and nutrient availability * Light is the main factor regulating plant development and physiology: Ø array of photoreceptors for optimal function in both different light intensities and qualities * two classes of photoreceptors for establishing period length 1) Phytochromes A-E * Phy. A main in dark-grown seedlings; rapidly degraded in light * Phy. B-E more light stable; phy. B main in light-grown seedlings 2) Cryptochromes 1 -2 (blue, UV-A region) maintenance of circadian period length under a whole range of light conditions

THE CENTRAL OSCILLATOR 1) CCA 1 and LHY * MYB-like DNA-binding domains of high similarity - TFs 2) TOC 1 * atypical response regulator (of His-kinase) *C-terminus similarity to the CONSTANS family of TF Ømediate pt-pt interactions and nuclear localization

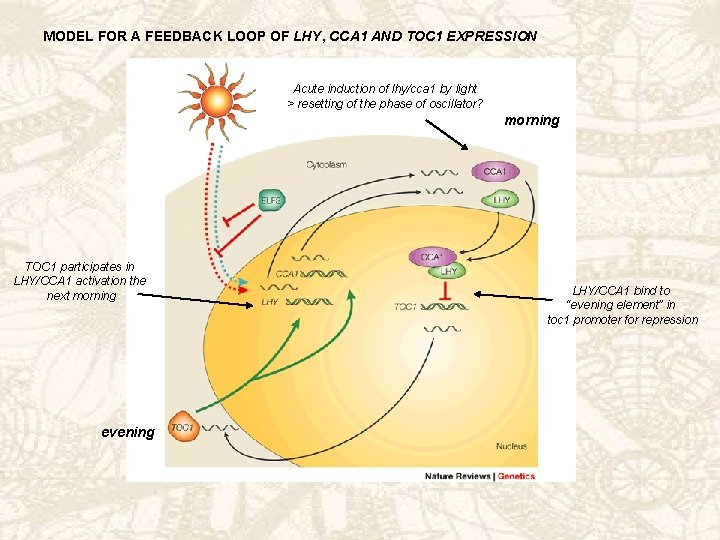
THE CENTRAL OSCILLATOR 1) CCA 1 and LHY * when overexpressed: > arrhythmicity under constant light or dark > reduction in m. RNA levels: negative feedback loop * loss of either gene affects periodicity but doesn’t abolish rhythmicity: overlapping functions * both m. RNA and PT levels oscillate, peaking at dawn 2) TOC 1 * mutations: period shortening independently of light, but still rhythmicity > other factors * model for a feedback loop involving LHY, CCA 1 and TOC 1 based on: a) Toc 1 expression oscillates peaking during early evening, opposite to CCA 1 and LHY b) TOC 1 expression low in LHY or CCA 1 overexpressors > transcriptional repression by CCA 1/LHY? c) TOC 1 expression high in lhy/cca 1 double mutants d) In TOC 1 mutants CCA 1/LHY expression very low > TOC 1 positive regulator of LHY/CCA 1?

MODEL FOR A FEEDBACK LOOP OF LHY, CCA 1 AND TOC 1 EXPRESSION Acute induction of lhy/cca 1 by light > resetting of the phase of oscillator? morning TOC 1 participates in LHY/CCA 1 activation the next morning evening LHY/CCA 1 bind to “evening element” in toc 1 promoter for repression

OTHER COMPONENTS a) PIF 3 * PAS-domain-containing b. HLH transcription factor * interacts with phy. B and binds to a number of phy-regulated promoters (e. g. CCA 1 and LHY) * TOC 1 binds PIF 3 > interaction necessary for LHY and CCA 1 activation? b) ELF 3 * Interacts with PHYB * arrhythmia only under constant light, while functional clock under constant darkness Ø not required for clock function in the absence of light * low [CCA 1 and LHY m. RNA] and high [TOC 1 m. RNA] * loss-of-function mutants Ø consistent with a negative role of CCA 1/LHY in TOC 1 regulation Ø ELF 3 required for TOC 1 promotion of lhy/cca 1 transcription? Ø elf 3 oscillates with the same phase as TOC 1 * clock hypersensitive to light in the night > causing arrhythmia * Seems to gate the light input to the oscillator, protecting it from the light signal at particular times of the day * ELF 3 might gate the light input at dusk so that the circadian clock is reset by the “light-on” signal

c) ZEITLUPE (ZTL) * long period for cab/other CCGs, which dependent on fluence rate > light input to the clock * interacts with Phy. B and CRY 1 * PAS domain + kelch domain for pt-pt interactions + F-box > targeting of pts to the proteasome? * Transcript levels don’t oscillate d) GIGANTEA (GI) * Shortens period of gene expression rhythms * less severe effect in darkness than light, and dependent on fluence rate > light input to the clock * Gi transcript levels oscillate * Involved in phy. B signaling * Some phenotypes as phy. B mutants (hypocotyl length) * Others are opposite: late flowering vs. early flowering phy. B mutants * Different roles in different developmental stages? * Branch of phy. B signaling?

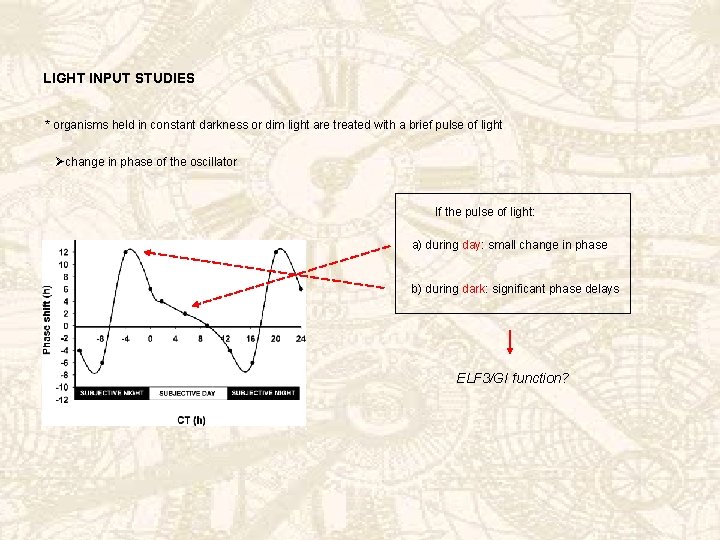
LIGHT INPUT STUDIES * organisms held in constant darkness or dim light are treated with a brief pulse of light Øchange in phase of the oscillator If the pulse of light: a) during day: small change in phase b) during dark: significant phase delays ELF 3/GI function?

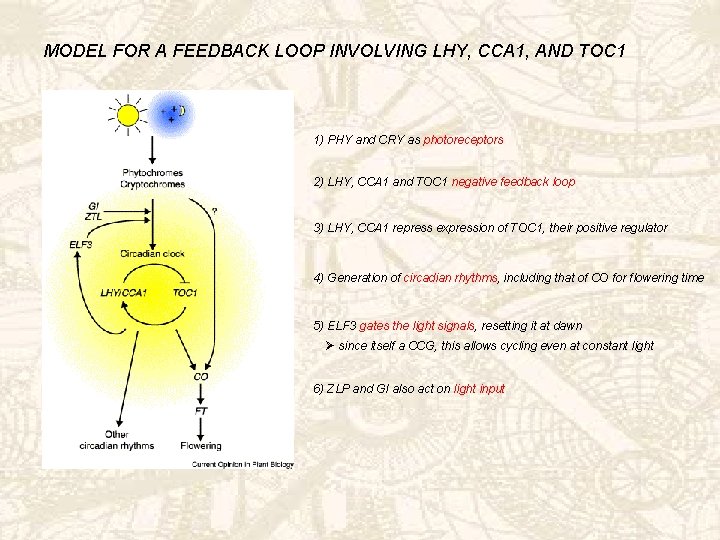
MODEL FOR A FEEDBACK LOOP INVOLVING LHY, CCA 1, AND TOC 1 1) PHY and CRY as photoreceptors 2) LHY, CCA 1 and TOC 1 negative feedback loop 3) LHY, CCA 1 repress expression of TOC 1, their positive regulator 4) Generation of circadian rhythms, including that of CO for flowering time 5) ELF 3 gates the light signals, resetting it at dawn Ø since itself a CCG, this allows cycling even at constant light 6) ZLP and GI also act on light input

SEASONAL RHYTHMS * In plants: formation of flowers at the most appropriate times of the year to ensure reproductive success * Controlled by changes in day length (photoperiodism) monitored by the circadian clock. * Normally, Arabidopsis flowers more rapidly in LD (summer) than SD conditions (winter) Ø misregulation or mutation of genes implicated in clock function disrupts this response * Daylength measurement is mediated by * transcriptional regulation of gene expression by the circadian clock, * post-transcriptional regulation by light * important information has been provided from several studies of the flowering-time gene CONSTANS Ø CO mutations cause delayed flowering under long days (LDs)

EVOLUTIONARY ASPECTS * No conservation of clock components among organisms > clocks have arisen multiple times e. g. cryptochromes implicated in clock function in many organisms arose independently from the DNA repair enzyme photolyase in an example of repeated evolution * Some common features: a) transcriptional-feedback loop that generates a circadian oscillation in the level of one or more critical clock components Ø this level defines the phase of the clock at any point b) presence of PAS protein domains (protein interactions) in critical clock components * mutations in single genes never eliminate rhythmicity > redundant functions * plants * more sophisticated light-entrainment strategy Øseveral photoreceptors that fine-tune the clock to different light conditions * primary driving force for clock evolution: "flight from light“, to set light-sensitive processes to occur at night Ø cell division in the alga Chlamydomonas occurs during the dark phase Ø genes encoding enzymes involved the synthesis of UV-protective compounds peak just before dawn in plants

MOLECULAR BASIS OF SEASONAL TIME MEASUREMENT IN ARABIDOPSIS (Yanovsky and Kay, Nature 2002) * Many signaling and clock components identified, but unknown how information integrated * CO isolated as a delayed flowering mutant under LDs * Expression of CO regulated by clock and photoperiod, with high CO during the day in LD but not in SD * CO promotes flowering through the induction of the flowering time gene FT * FT peaks when CO expression is high and illumination > so CO activity regulated by light? * In gi, lhy and elf 3 mutants CO levels correlate with flowering time alterations Ø mutants affected also in light signaling > difficult to distinguish between circadian and light effect

* How does CO promote flowering through FT? * How to distinguish between the circadian and light effect? * toc 1 mutant chosen for approaching these questions Ø pseudo response regulator closely associated with the central oscillator Ø when mutated: period shortening of all studied output genes even in complete darkness Ø early flowering under SD of 24 h Ø normal flowering under SD of 21 h, that match the endogenous period of the mutant

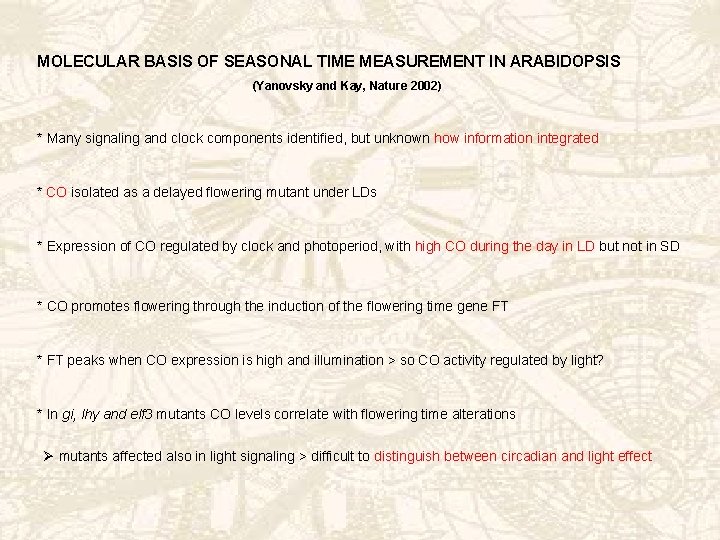
1. CO-EXPRESSION IN TOC 1 VS. WT PLANTS SD LD WT SD (24 and 21): CO accumulation during dark LD: CO accumulation also during dawn and dusk toc 1 SD 24: CO accumulation advanced: high during dusk – early flowering No changes in level of expression or curve shape SD 21 and LD: CO expression – and flowering like WT

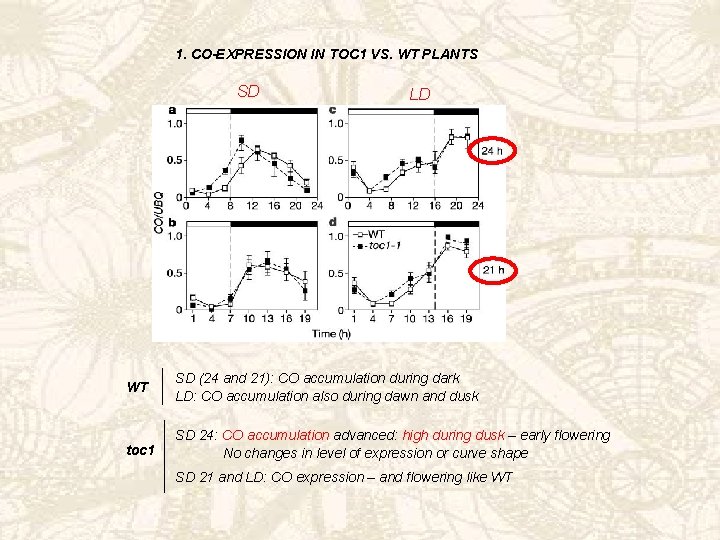
Co accelerates flowering through direct activation of FT expression: same mechanism here? 2. FT-EXPRESSION IN TOC 1 VS. WT PLANTS SD WT LD SD (24 and 21): no FT accumulation LD: FT accumulation during illumination SD 24: FT accumulation during illumination toc 1 SD 21 and LD: FT expression like WT

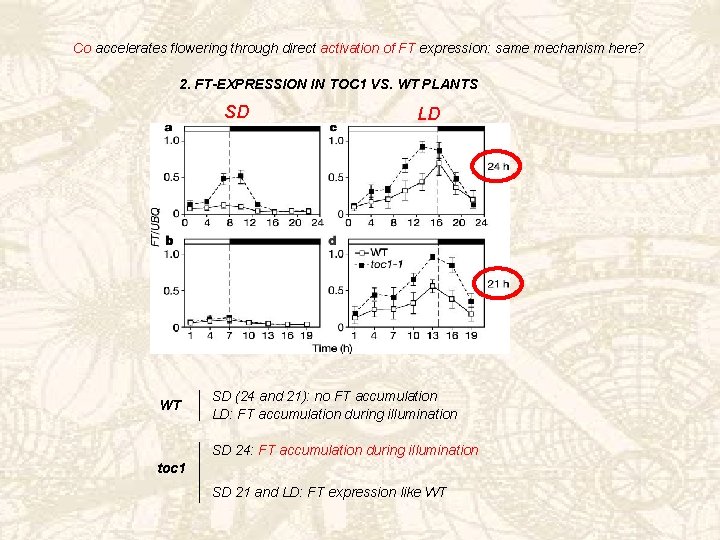
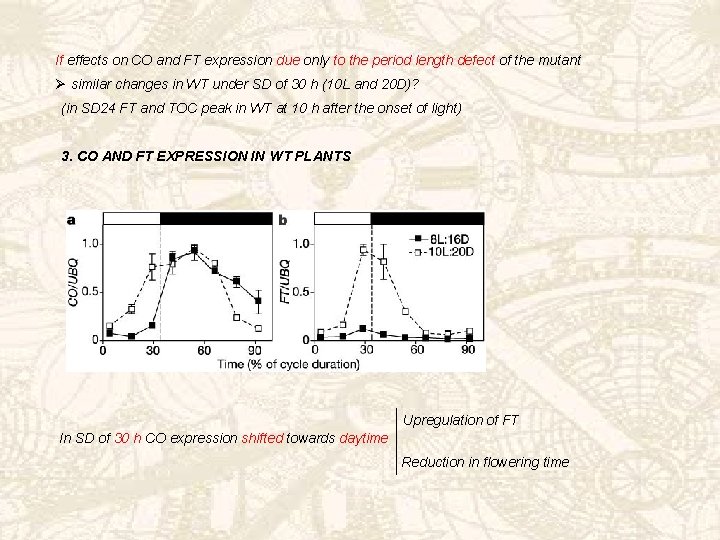
If effects on CO and FT expression due only to the period length defect of the mutant Ø similar changes in WT under SD of 30 h (10 L and 20 D)? (in SD 24 FT and TOC peak in WT at 10 h after the onset of light) 3. CO AND FT EXPRESSION IN WT PLANTS Upregulation of FT In SD of 30 h CO expression shifted towards daytime Reduction in flowering time

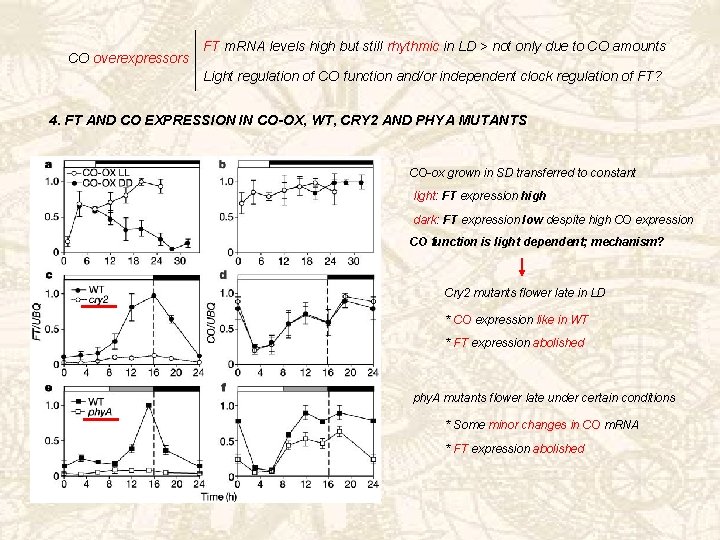
CO overexpressors FT m. RNA levels high but still rhythmic in LD > not only due to CO amounts Light regulation of CO function and/or independent clock regulation of FT? 4. FT AND CO EXPRESSION IN CO-OX, WT, CRY 2 AND PHYA MUTANTS CO-ox grown in SD transferred to constant light: FT expression high dark: FT expression low despite high CO expression CO function is light dependent; mechanism? Cry 2 mutants flower late in LD * CO expression like in WT * FT expression abolished phy. A mutants flower late under certain conditions * Some minor changes in CO m. RNA * FT expression abolished

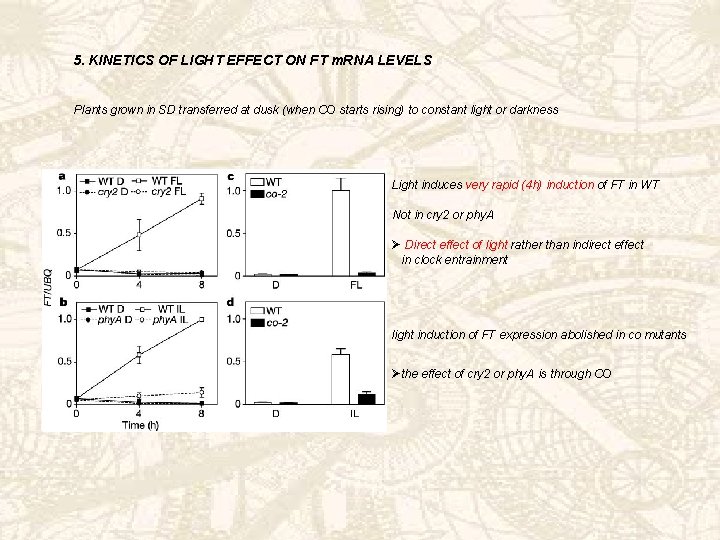
5. KINETICS OF LIGHT EFFECT ON FT m. RNA LEVELS Plants grown in SD transferred at dusk (when CO starts rising) to constant light or darkness Light induces very rapid (4 h) induction of FT in WT Not in cry 2 or phy. A Ø Direct effect of light rather than indirect effect in clock entrainment light induction of FT expression abolished in co mutants Øthe effect of cry 2 or phy. A is through CO

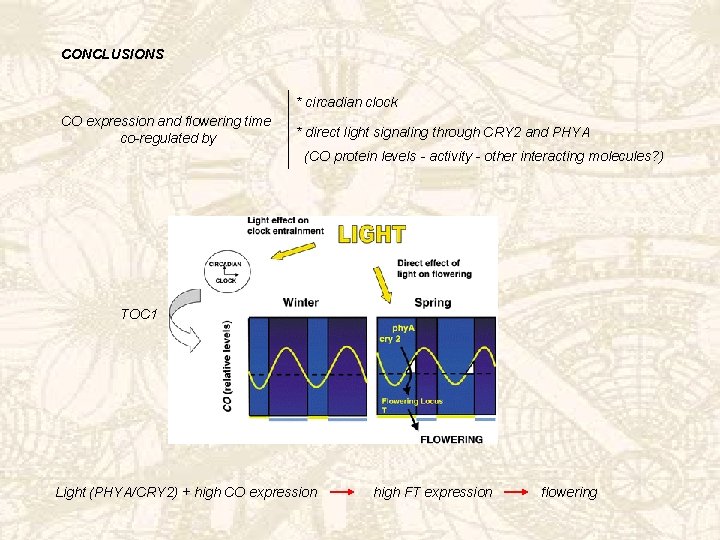
CONCLUSIONS * circadian clock CO expression and flowering time co-regulated by * direct light signaling through CRY 2 and PHYA (CO protein levels - activity - other interacting molecules? ) TOC 1 Light (PHYA/CRY 2) + high CO expression high FT expression flowering

The Arabidopsis SRR 1 gene mediates phy. B signaling and is required for normal circadian clock function (Staiger et al. 2003, Gen. Dev. ) * Light input to the oscillator through phytochromes and cryptochromes Activates phytochrome through conformational changes * Light Affects phytochrome subcellular localization Affects their ability to interact with signaling partners * Many signaling components downstream of PHY Ø best characterised: interaction of phy. B with PIF 3 to activate e. g. CCA 1 * Here: srr 1(sensitivity to red light reduced) a new Arabidopsis mutant altered in * multiple outputs of the clock * phy. B light signaling

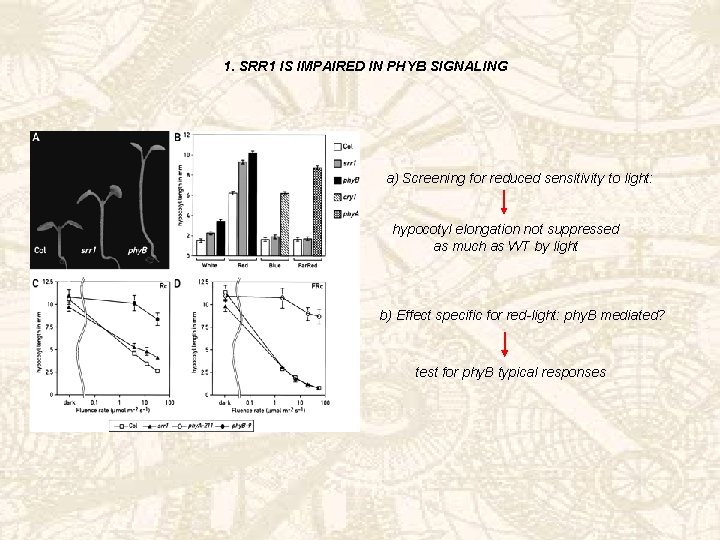
1. SRR 1 IS IMPAIRED IN PHYB SIGNALING a) Screening for reduced sensitivity to light: hypocotyl elongation not suppressed as much as WT by light b) Effect specific for red-light: phy. B mediated? test for phy. B typical responses

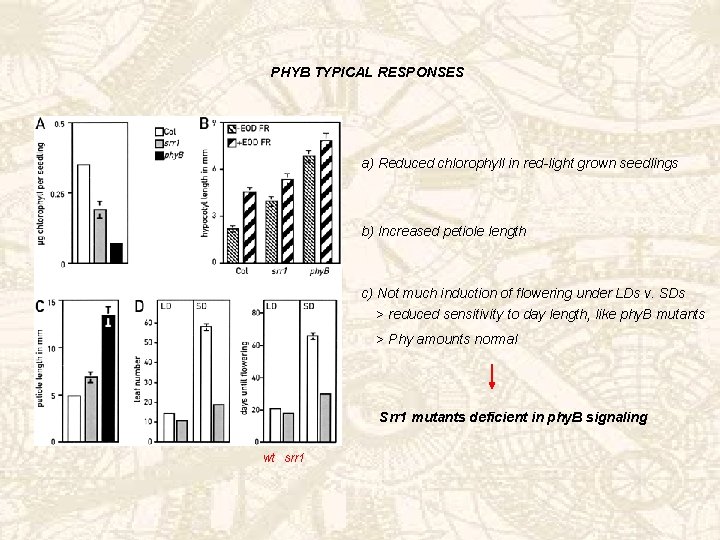
PHYB TYPICAL RESPONSES a) Reduced chlorophyll in red-light grown seedlings b) Increased petiole length c) Not much induction of flowering under LDs v. SDs > reduced sensitivity to day length, like phy. B mutants > Phy amounts normal Srr 1 mutants deficient in phy. B signaling wt srr 1

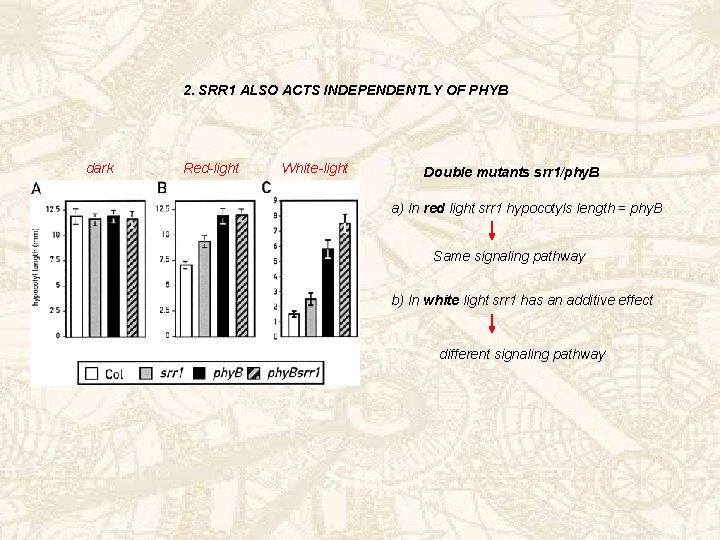
2. SRR 1 ALSO ACTS INDEPENDENTLY OF PHYB dark Red-light White-light Double mutants srr 1/phy. B a) In red light srr 1 hypocotyls length = phy. B Same signaling pathway b) In white light srr 1 has an additive effect different signaling pathway

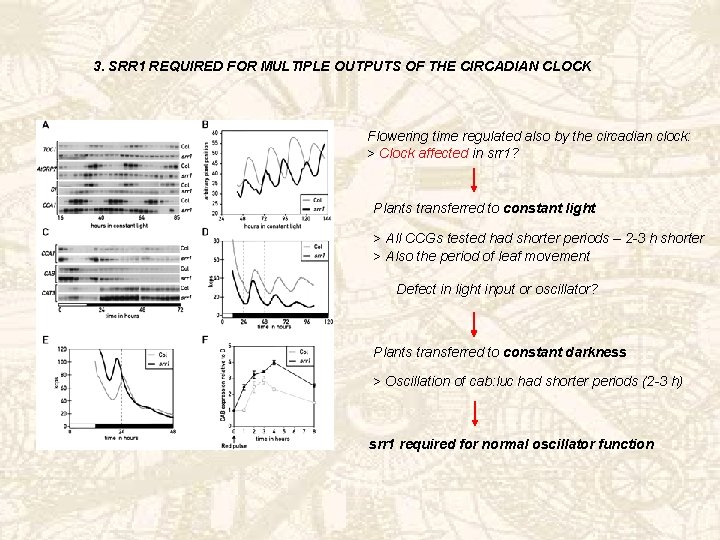
3. SRR 1 REQUIRED FOR MULTIPLE OUTPUTS OF THE CIRCADIAN CLOCK Flowering time regulated also by the circadian clock: > Clock affected in srr 1? Plants transferred to constant light > All CCGs tested had shorter periods – 2 -3 h shorter > Also the period of leaf movement Defect in light input or oscillator? Plants transferred to constant darkness > Oscillation of cab: luc had shorter periods (2 -3 h) srr 1 required for normal oscillator function

4. SRR 1: A CONSERVED NUCLEAR/CYTOPLASMIC PROTEIN * no recognizable domains * Putative nuclear localization sequence ØSRR 1 -GFP fusion: SRR 1 found both in cytoplasm and nucleus 5. THE SRR 1 TRANSCRIPT IS INDUCED BY LIGHT * m. RNA levels constant across the circadian cycle * Induced by red but not far-red light – phy. B signaling

6. CONCLUSIONS * srr 1: role in phy. B signaling and in regulation of circadian clock (like ELF 3 and GI) * elf 3: arrhythmia in light but remains rhythmic in darkness > light input to clock * srr 1 circadian phenotype both in light and darknes > required for normal oscillator function * elf 3 interacts with phy. B in vitro > Interaction between srr 1 and phy. B? * srr 1 homolog even in human * Lack of conservation among clock components, but some common features Ø srr 1 involved in some of these? (regulated nuclear translocation, phosphorylation and negative feedback loops)