Chromosomes Structure and Function MOLECULAR GENETICS 856 Chromosomes








- Slides: 46

Chromosomes: Structure and Function MOLECULAR GENETICS (856)

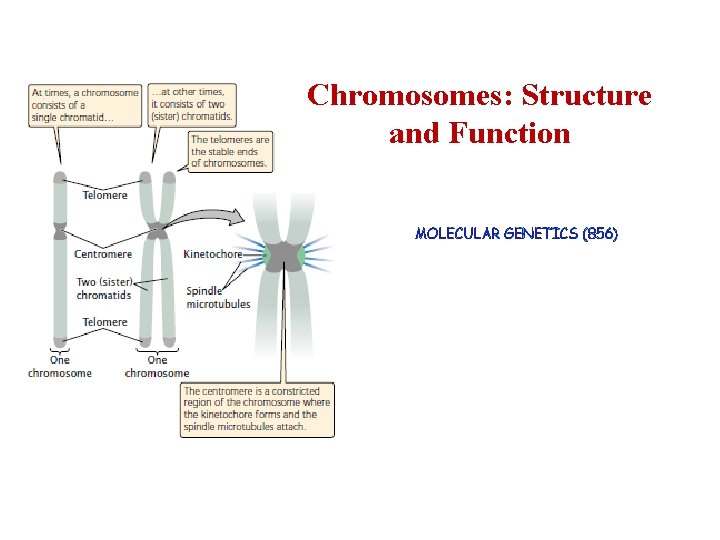
Chromosomes • Transmission • Expression of genetic information. • A centromere, which is most evident at metaphase, where it is the narrowest part of the chromosome and the region at which spindle fibers attach. • Replication origins, certain DNA sequences along each chromosome at which DNA replication can be initiated. • Telomeres, the ends of linear chromosomes that have a specialized structure to prevent internal DNA from being degraded by nucleases.

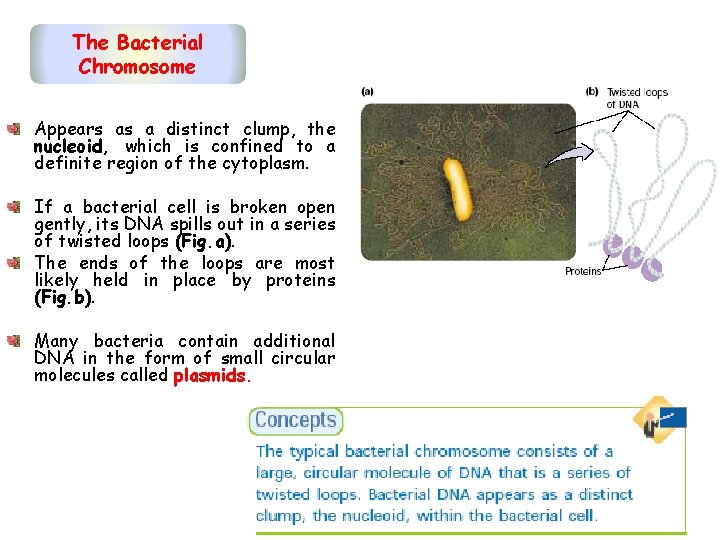
The Bacterial Chromosome Appears as a distinct clump, the nucleoid, which is confined to a definite region of the cytoplasm. If a bacterial cell is broken open gently, its DNA spills out in a series of twisted loops (Fig. a). The ends of the loops are most likely held in place by proteins (Fig. b). Many bacteria contain additional DNA in the form of small circular molecules called plasmids.

The Eukaryotic Chromosome Individual eukaryotic chromosomes contain enormous amounts of DNA, and consists of a single, extremely long molecule of DNA. The chromosomes are in an elongated, relatively uncondensed state (less tightly packed than DNA in mitotic chromosomes) during interphase of the cell cycle.

The Eukaryotic Chromosome In the course of the cell cycle, the level of DNA packaging changes— chromosomes progress from a highly packed state to a state of extreme condensation. DNA packaging also changes locally in replication and transcription, when the two nucleotide strands must unwind so that particular base sequences are exposed. Thus, the packaging of eukaryotic DNA is not static but changes regularly in response to cellular processes.

Packing DNA into Small Spaces Ø Chromosome of the bacterium E. coli, a single molecule of DNA with approximately 4. 64 million base pairs. Stretched out straight, this DNA would be about 1000 times as long as the cell within which it resides. Ø Human cells contain 6 billion base pairs of DNA, which would measure some 1. 8 meters stretched end to end. Ø Even DNA in the smallest human chromosome would stretch 14, 000 times the length of the nucleus. DNA molecules must be tightly packed to fit into such small spaces.

Chromosomal DNA is coiled hierarchically The structure of DNA can be considered at three hierarchical levels: the primary structure of DNA is its nucleotide sequence; the secondary structure is the double stranded helix; and the tertiary structure refers to higher order folding that allows DNA to be packed into the confined space of a cell.

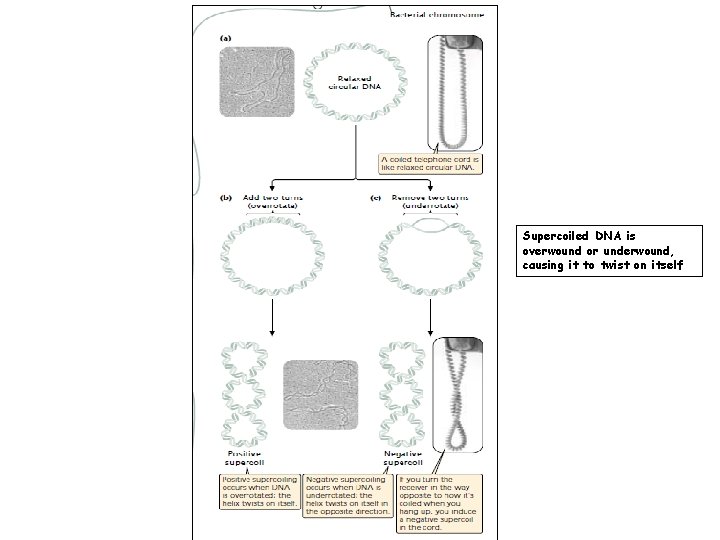
DNA Supercoils Most DNA found in cells is negatively supercoiled, which has two advantages for the cell. – First, supercoiling makes the separation of the two strands of DNA easier during replication and transcription. Negatively supercoiled DNA is underrotated; so separation of the two strands during replication and transcription is more rapid and requires less energy. – Second, supercoiled DNA can be packed into a smaller space because it occupies less volume than relaxed DNA.

Supercoiled DNA is overwound or underwound, causing it to twist on itself

Supercoiling; Tertiary Structures Ø Supercoiling occurs Ø only if the two polynucleotide strands of the DNA double helix are unable to rotate about each other freely. If the chains can turn freely, their ends will simply turn as extra rotations are added or removed, and the molecule will spontaneously revert to the relaxed state. Ø When the strain of over rotating or under rotating cannot be compensated by the turning of the ends of the double helix, which is the case if the DNA is circular— that is, there are no free ends. Some viral chromosomes are in the form of simple circles and readily undergo supercoiling. Ø Eukaryotic DNA is normally linear but also tends to fold into loops stabilized by proteins. In these chromosomes, the anchoring proteins prevent free rotation of the ends of the DNA; so super coiling does take place.

DNA Supercoils: Enzymes v Topoisomerases, enzymes that add or remove rotations from the DNA helix by temporarily breaking the nucleotide strands, rotating the ends around each other, and then rejoining the broken ends. v The two classes of topoisomerases are: v Type I, which breaks only one of the nucleotide strands and reduces super coiling by removing rotations. v Type II, which adds or removes rotations by breaking both nucleotide strands

Chromatin Structure Eukaryotic DNA is closely associated with proteins, creating chromatin. Two basic types of chromatin are: o Euchromatin, which undergoes the normal process of condensation and decondensation in the cell cycle o Heterochromatin, which remains in a highly condensed state throughout the cell cycle, even during interphase. Proteins in chromatin are the histones, which are relatively small, positively charged proteins of five major types: H 1, H 2 A, H 2 B, H 3, and H 4. All histones have a high percentage of arginine and lysine, positively charged amino acids that give them a net positive charge. The positive charges attract the negative charges on the phosphates of DNA and holds the DNA in contact with the histones.

Euchromatin Exist in extended state, dispersed through the nucleus and staining diffusely. Early-replicating and GC rich region. In prokaryotes, euchromatin is the only form of chromatin present. Genes may oy may not expressed ÄHeterochromatin Ädarkly stained 1. Constitutive ÄConsist ÄLate ; always inactive and condensed. of repetitive DNA replicating and AT rich region ÄPresent Ä two types at identical positions on all chromosomes in all cell types of an organism. Genes poorly expressed. Human chromosomes 1, 9, 16, and the Y chromosome contain large regions of constitutive heterochromatin. Ä ÄOccurs around the centromere and near telomeres. 2. Facultative; Genetically active(decondensed) and inactive (condensed) Ä Variable in its expression. It varies with the cell type and may be manifested as condensed, or heavily stained.

Chromatin Structure Non-histone chromosomal proteins make up about half of the protein mass of the chromosome. Non-histone chromosomal proteins serves structural roles and those that take part in genetic processes such as transcription and replication. Chromosomal scaffold proteins are revealed when chromatin is treated with a concentrated salt solution, which removes histones and most other chromosomal proteins, leaving a chromosomal protein “skeleton” to which the DNA is attached. These scaffold proteins may play a role in the folding and packing of the chromosome. Other structural proteins make up the kinetochore, cap the chromosome ends by attaching to telomeres, and constitute the molecular motors that move chromosomes in mitosis and meiosis.

Chromatin Structure • Other types of nonhistone chromosomal proteins play a role in genetic processes. – components of the replication machinery (DNA polymerases, helicases, primases and proteins that carry out and regulate transcription (RNA polymerases, transcription factors, acetylases). • High-mobility-group proteins are small, highly charged proteins that vary in amount and composition, depending on tissue type and stage of the cell cycle. – these proteins may play an important role in altering the packing of chromatin during transcription.

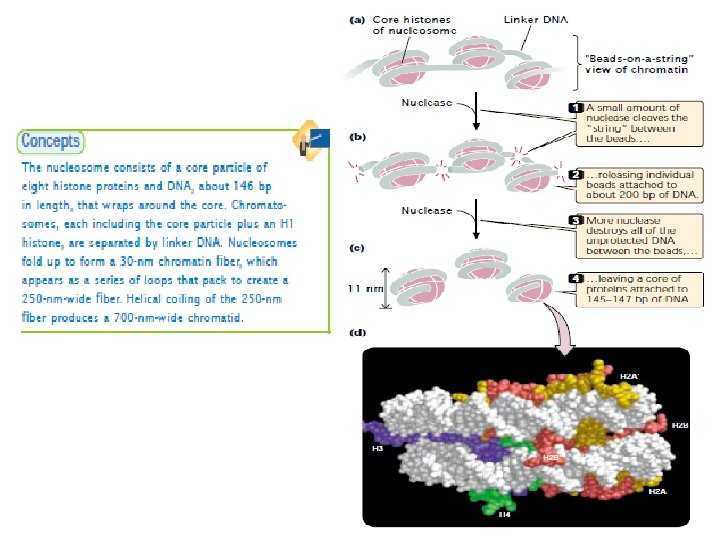
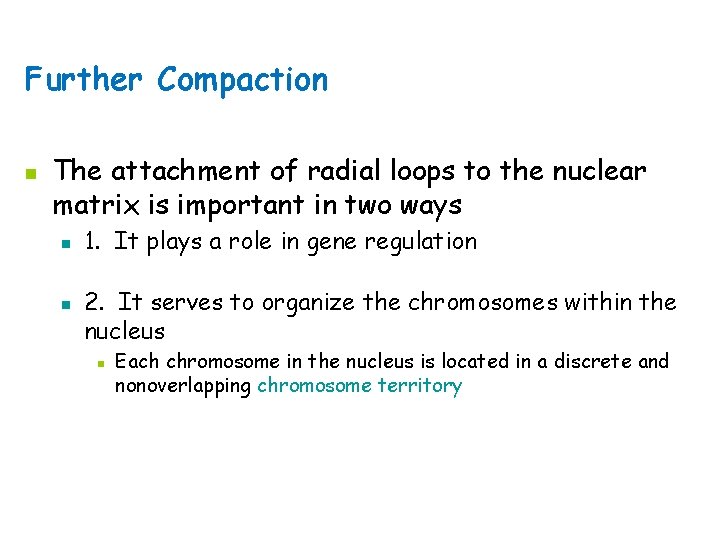
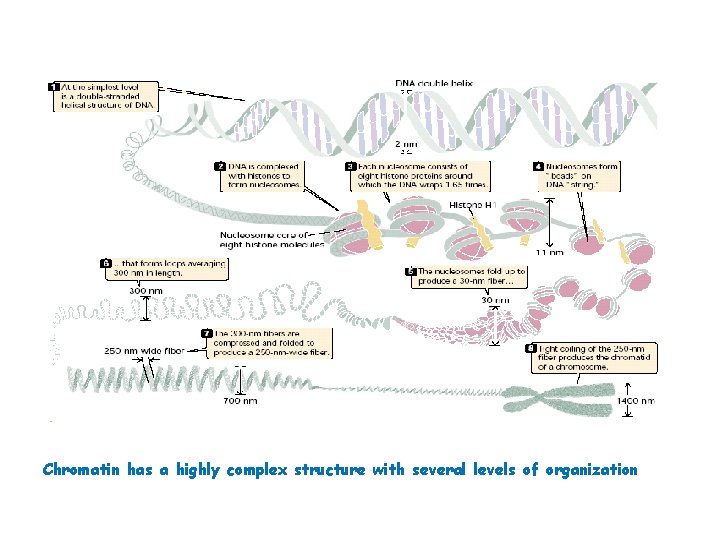
Nucleosome Chromatin has a highly complex structure with several levels of organization. The simplest level is the double helical structure of DNA. At a more complex level, the DNA molecule is associated with proteins and is highly folded to produce a chromosome. When chromatin is isolated from the nucleus of a cell and viewed with an electron microscope, it frequently looks like beads on a string (Fig. a), If a small amount of nuclease is added to this structure, the enzyme cleaves the string between the beads, leaving individual beads attached to about 200 bp of DNA (Fig. b). If more nuclease is added, the enzyme chews up all of the DNA between the beads and leaves a core of proteins attached to a fragment of DNA (Fig. c). The repeating core of protein and DNA produced by digestion with nuclease enzymes is the simplest level of chromatin structure, the Nucleosome. The nucleosome is a core particle consisting of DNA wrapped about two times around an octamer of eight histone proteins (two copies each of H 2 A, H 2 B, H 3, and H 4) (Fig. d). The DNA in direct contact with the histone octamer is between 145 and 147 bp in length, coils around the histones in a left-handed direction, and is supercoiled. It does not wrap around the octamer smoothly; there are four bends in its helical structure as it winds around the histones.


Nucleosome The fifth type of histone, H 1, is not a part of the core particle but plays an important role in the nucleosome structure. The precise location of H 1 with respect to the core particle is still uncertain. H 1 sits outside the octamer and binds to the DNA where the DNA joins and leaves the octamer. However, the results of recent experiments suggest that the H 1 histone sits inside the coils of the nucleosome. H 1 helps to lock the DNA into place, acting as a clamp around the nucleosome octamer. Together, the core particle and its associated H 1 histone are called the chromatosome, the next level of chromatin organization. The H 1 protein is attached to between 20 and 22 bp of DNA, and the nucleosome encompasses an additional 145 to 147 bp of DNA; so about 167 bp of DNA are held within the chromatosome. Chromatosomes are located at regular intervals along the DNA molecule and are separated from one another by linker DNA, which varies in size among cell types—most cells have from about 30 bp to 40 bp of linker DNA. Non-histone chromosomal proteins may be associated with this linker DNA, and a few also appear to bind directly to the core particle.

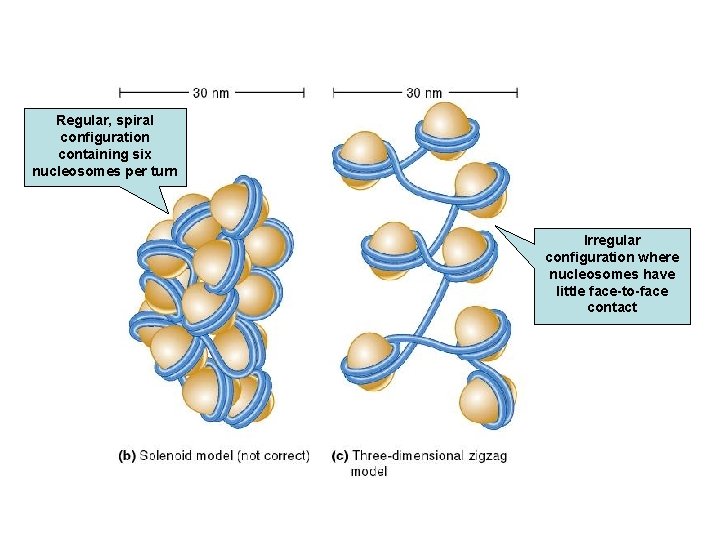
Nucleosomes Join to Form a 30 nm Fiber n Nucleosomes associate with each other to form a more compact structure termed the 30 nm fiber n Histone H 1 plays a role in this compaction n At moderate salt concentrations, H 1 is removed n n The result is the classic beads-on-a-string morphology At low salt concentrations, H 1 remains bound n Beads associate together into a more compact morphology

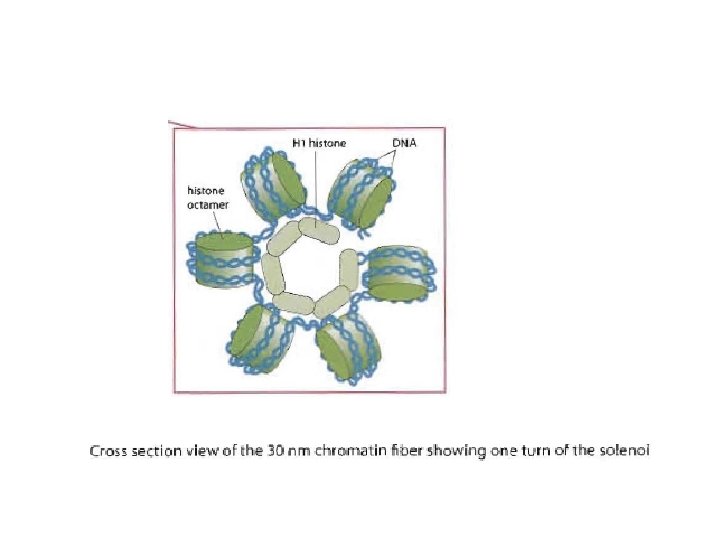
n n The 30 nm fiber shortens the total length of DNA another seven-fold Its structure has proven difficult to determine n n The DNA conformation may be substantially altered when extracted from living cells Two models : n n Solenoid model Three-dimensional zigzag model

Regular, spiral configuration containing six nucleosomes per turn Irregular configuration where nucleosomes have little face-to-face contact

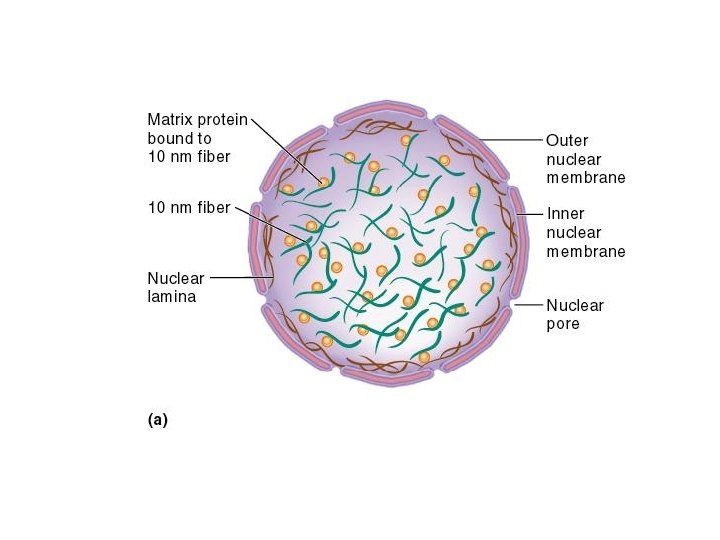
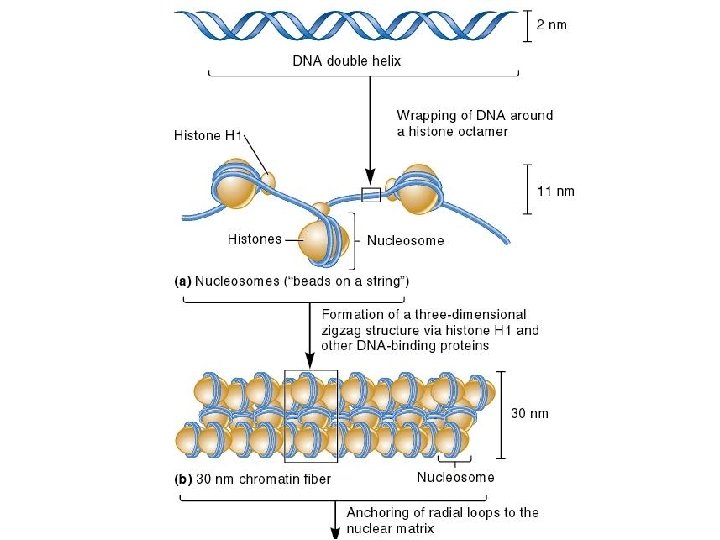
Further Compaction of the Chromosome n n n The two events we have discussed so far have shortened the DNA about 50 -fold A third level of compaction involves interaction between the 30 nm fiber and the nuclear matrix The nuclear matrix is composed of two parts n Nuclear lamina n Internal matrix proteins n 10 nm fiber and associated proteins


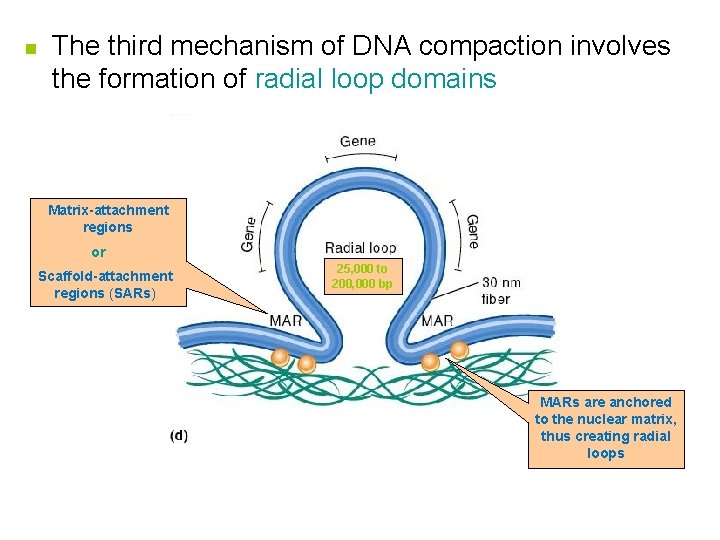
n The third mechanism of DNA compaction involves the formation of radial loop domains Matrix-attachment regions or Scaffold-attachment regions (SARs) 25, 000 to 200, 000 bp MARs are anchored to the nuclear matrix, thus creating radial loops

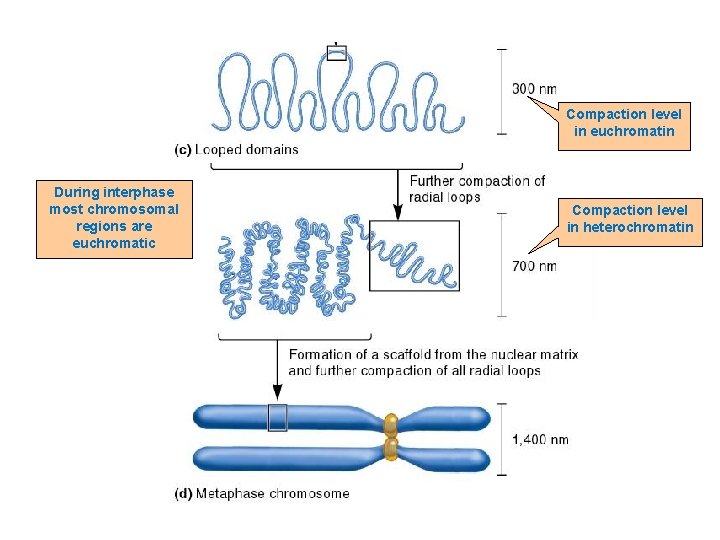
Further Compaction n The attachment of radial loops to the nuclear matrix is important in two ways n n 1. It plays a role in gene regulation 2. It serves to organize the chromosomes within the nucleus n Each chromosome in the nucleus is located in a discrete and nonoverlapping chromosome territory


Compaction level in euchromatin During interphase most chromosomal regions are euchromatic Compaction level in heterochromatin


Chromatin has a highly complex structure with several levels of organization

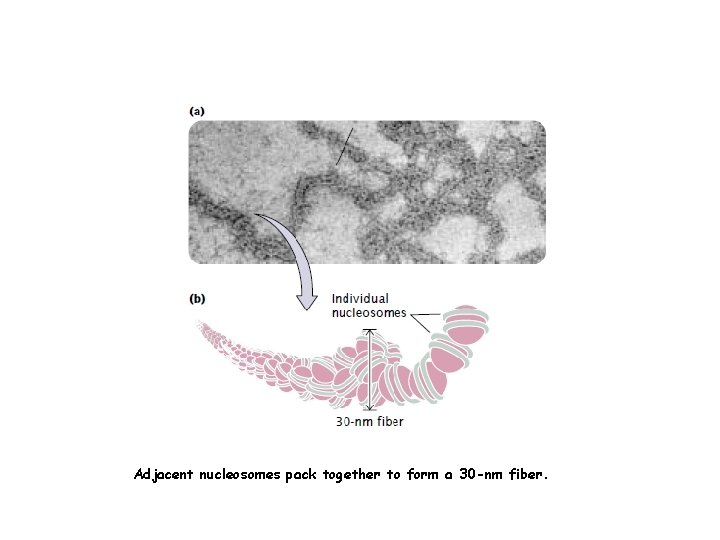
Higher-order chromatin structure In chromosomes, adjacent nucleosomes are not separated by space equal to the length of the linker DNA; rather, nucleosomes fold on themselves to form a dense, tightly packed structure. This structure is revealed when nuclei are gently broken, open and their contents are examined with the use of an electron microscope; much of the chromatin that spills out appears as a fiber with a diameter of about 30 nm (Fig a). The next-higher level of chromatin structure is a series of loops of 30 -nm fibers, each anchored at its base by proteins in the nuclear scaffold. On average, each loop encompasses some 20, 000 to 100, 000 bp of DNA and is about 300 nm in length, but the individual loops vary considerably. The 300 -nm fibers are packed and folded to produce a 250 -nm-wide fiber. Tight helical coiling of the 250 -nm fiber, in turn, produces the structure that appears in metaphase: an individual chromatid approximately 700 nm in width.

Adjacent nucleosomes pack together to form a 30 -nm fiber.

Changes in Chromatin Structure Eukaryotic DNA must be tightly packed to fit into the cell nucleus, it must also periodically unwind to undergo replication and transcription. Evidence of the changing nature of chromatin structure is seen in the puffs of polytene chromosomes and in the sensitivity of genes to digestion by DNase I. Polytene chromosomes are giant chromosomes found in certain tissues of Drosophila and some other organisms. These large, unusual chromosomes arise when repeated rounds of DNA replication take place without accompanying cell divisions, producing thousands of copies of DNA that lie side by side. When polytene chromosomes are stained with dyes, numerous bands are revealed. Under certain conditions, the bands may exhibit chromosomal puffs—localized swellings of the chromosome. Each puff is a region of the chromatin that has relaxed its structure, assuming a more open state. If radioactively labeled uridine is added to a Drosophila larva, radioactivity accumulates in chromosomal puffs, indicating that they are regions of active transcription. Appearance of puffs at particular locations on the chromosome can be stimulated by exposure to hormones and other compounds that are known to induce the transcription of genes at those locations. This correlation between the occurrence of transcription and the relaxation of chromatin at a puff site indicates that chromatin structure undergoes dynamic change associated with gene activity.

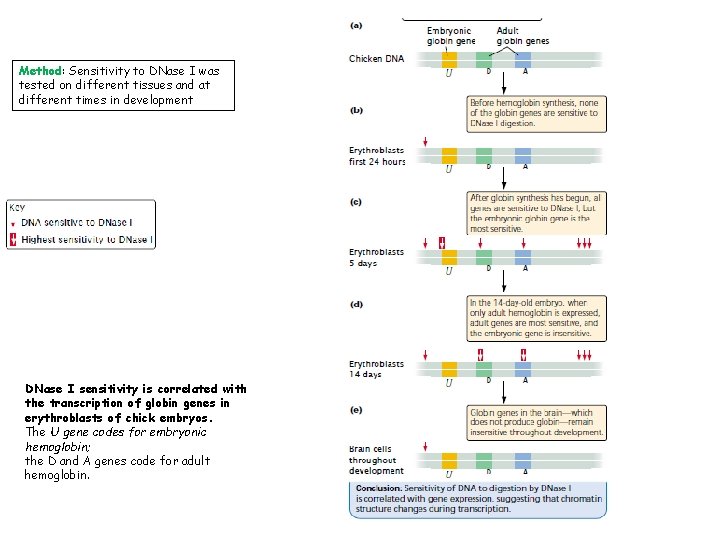
A several experiments indicating that chromatin structure changes with gene activity is sensitivity to DNase I, an enzyme that digests DNA. The ability of this enzyme to digest DNA depends on chromatin structure: when DNA is tightly bound to histone proteins, it is less sensitive to DNase I, whereas unbound DNA is more sensitive to digestion by DNase I. The results of experiments that examine the effect of DNase I on specific genes show that DNase sensitivity is correlated with gene activity. ü For example, globin genes code for hemoglobin in the erythroblasts (precursors of red blood cells) of chickens. The forms of hemoglobin produced in chick embryos and chickens are different and are encoded by different genes (Fig. a). ü No hemoglobin is synthesized in chick embryos in the first 24 hours after fertilization. If DNase I is applied to chromatin from chick erythroblasts in this first 24 -hour period, all the globin genes are insensitive to digestion (Fig. b).

Changes in Chromatin Structure ü From day 2 to day 6 after fertilization, after hemoglobin synthesis has begun, the globin genes become sensitive to DNase I, and the genes that code for embryonic hemoglobin are the most sensitive (Fig c). ü After 14 days of development, embryonic hemoglobin is replaced by the adult forms of hemoglobin. The most sensitive regions now lie near the genes that produce the adult hemoglobins (Fig. d). DNA from brain cells, which produce no hemoglobin, remains insensitive to DNase digestion throughout development (Fig. e).

Method: Sensitivity to DNase I was tested on different tissues and at different times in development DNase I sensitivity is correlated with the transcription of globin genes in erythroblasts of chick embryos. The U gene codes for embryonic hemoglobin; the D and A genes code for adult hemoglobin.

Changes in Chromatin Structure In summary, when genes become transcriptionally active, they also become sensitive to DNase I, indicating that the chromatin structure is more exposed during transcription. What is the nature of the change in chromatin structure that produces chromosome puffs and DNase I sensitivity? In both cases, the chromatin relaxes; histones loosen their grip on the DNA. One process that appears to be implicated in changing chromatin structure is acetylation, a reaction that adds chemical groups called acetyls to the histone proteins. Enzymes called acetyltransferases attach acetyl groups to lysine amino acids at one end (called a tail) of the histone protein. This modification reduces the positive charges that normally exist on lysine and destabilizes the nucleosome structure, and so the histones hold the DNA less tightly. Proteins taking part in transcription can then bind more easily to the DNA and carry out transcription.

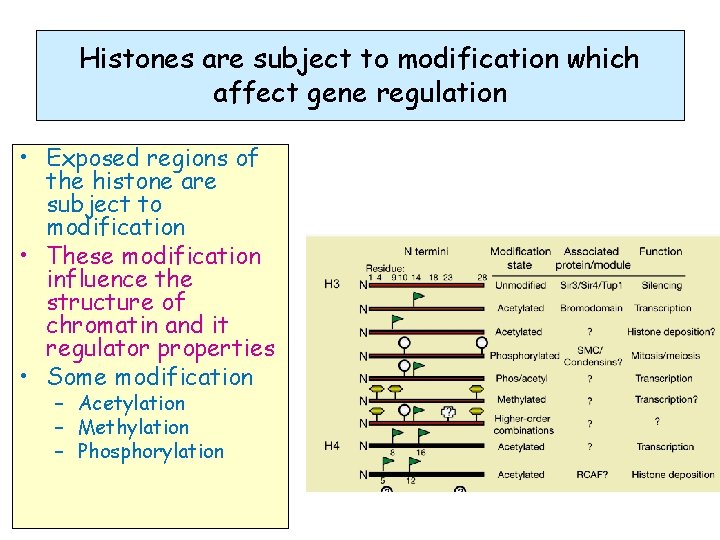
Histones are subject to modification which affect gene regulation • Exposed regions of the histone are subject to modification • These modification influence the structure of chromatin and it regulator properties • Some modification – Acetylation – Methylation – Phosphorylation

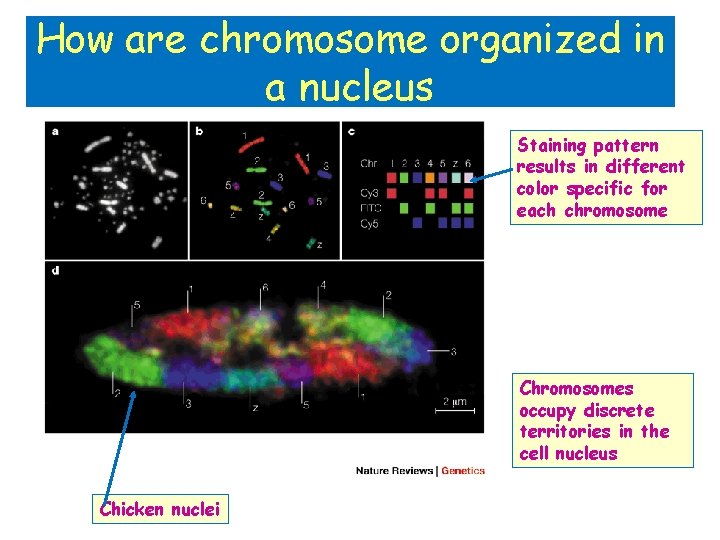
How are chromosome organized in a nucleus Staining pattern results in different color specific for each chromosome Chromosomes occupy discrete territories in the cell nucleus Chicken nuclei

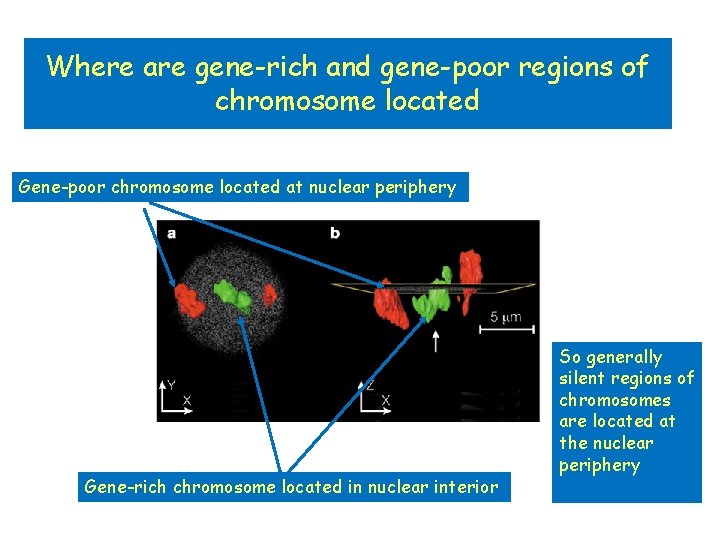
Where are gene-rich and gene-poor regions of chromosome located Gene-poor chromosome located at nuclear periphery Gene-rich chromosome located in nuclear interior So generally silent regions of chromosomes are located at the nuclear periphery

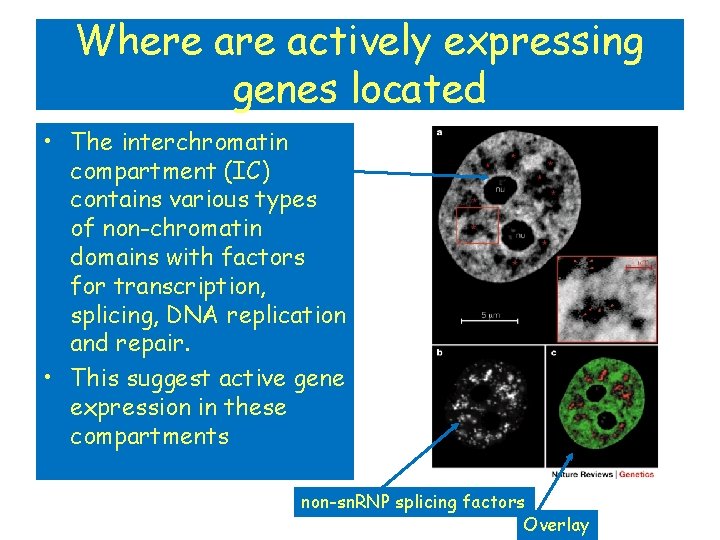
Where actively expressing genes located • The interchromatin compartment (IC) contains various types of non-chromatin domains with factors for transcription, splicing, DNA replication and repair. • This suggest active gene expression in these compartments non-sn. RNP splicing factors Overlay

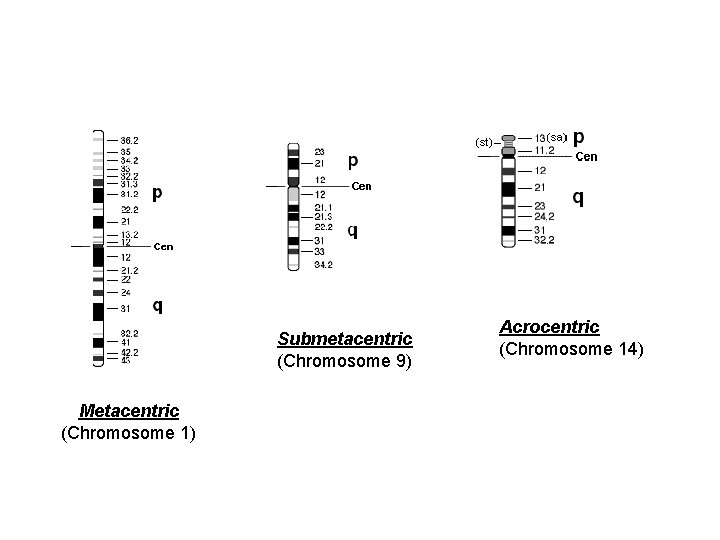
Chromosome Morphology Under the microscope chromosomes appear as thin, thread-like structures. • • They all have a short arm and long arm separated by a primary constriction called the centromere. The short arm is designated as p and the long arm as q. The centromere is the location of spindle attachment and is an integral part of the chromosome. It is essential for the normal movement and segregation of chromosomes during cell division. Human metaphase chromosomes can be categorized according to the length of the short and long arms and also the centromere location. • Metacentric chromosomes have short and long arms of roughly equal length with the • • centromere in the middle. Submetacentric chromosomes have short and long arms of unequal length with the centromere more towards one end. Acrocentric chromosomes have a centromere very near to one end and have very small short arms. They frequently have secondary constrictions on the short arms that connect very small pieces of DNA, called stalks and satellites, to the centromere. The stalks contain genes which code for ribosomal RNA. The diagrams showing region on chromosomes, called ideograms.

Submetacentric (Chromosome 9) Metacentric (Chromosome 1) Acrocentric (Chromosome 14)

• The ideogram is basically a "chromosome map" showing the relationship between the short and long arms, centromere (cen), and in the case of acrocentric chromosomes the stalks (st) and satellites (sa). Each band is numbered to aid in describing rearrangements.

Chromosome Nomenclature q The International System for Human Cytogenetic Nomenclature (ISCN) is fixed by the Standing Committee on Human Cytogenetic Nomenclature (Mitelman, 1995). q Basic terminology for banded chromosomes was decided at a meeting in Paris in 1971, and is often referred to as the Paris nomenclature. Ø Short arm locations are labeled p (petit) Ø Long arms are labelled q (queue). q Each chromosome arm is divided into regions labeled p 1, p 2, p 3 etc. , and q 1, q 2, q 3, etc. , counting outwards from the centromere. q Regions are delimited by specific landmarks, which are consistent and distinct morphological features, such as the ends of the chromosome arms, the centromere and certain bands. q Regions are divided into bands labeled p 11 (one-one, not eleven), p 12, p 13, etc. , q sub-bands labeled p 11. 1, p 11. 2 etc. , and q sub-bands e. g. p 11. 21, p 11. 22, etc. , in each case counting outwards from the centromere.

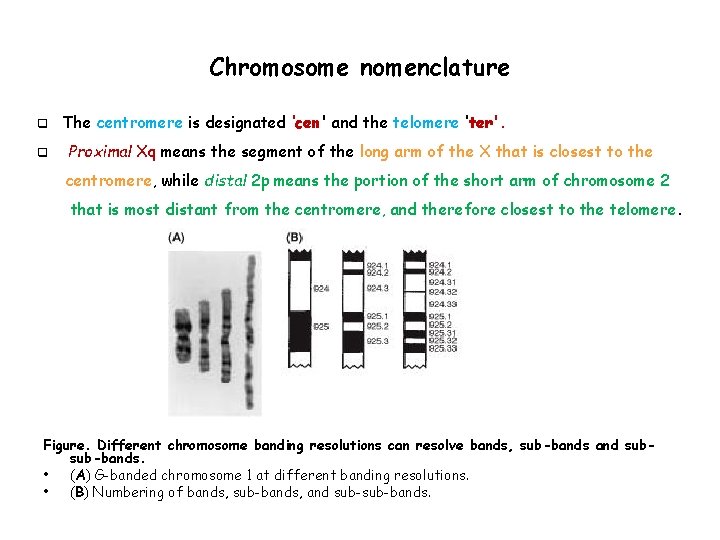
Chromosome nomenclature q q The centromere is designated ‘cen' and the telomere ‘ter'. Proximal Xq means the segment of the long arm of the X that is closest to the centromere, while distal 2 p means the portion of the short arm of chromosome 2 that is most distant from the centromere, and therefore closest to the telomere. Figure. Different chromosome banding resolutions can resolve bands, sub-bands and subsub-bands. • (A) G-banded chromosome 1 at different banding resolutions. • (B) Numbering of bands, sub-bands, and sub-bands.

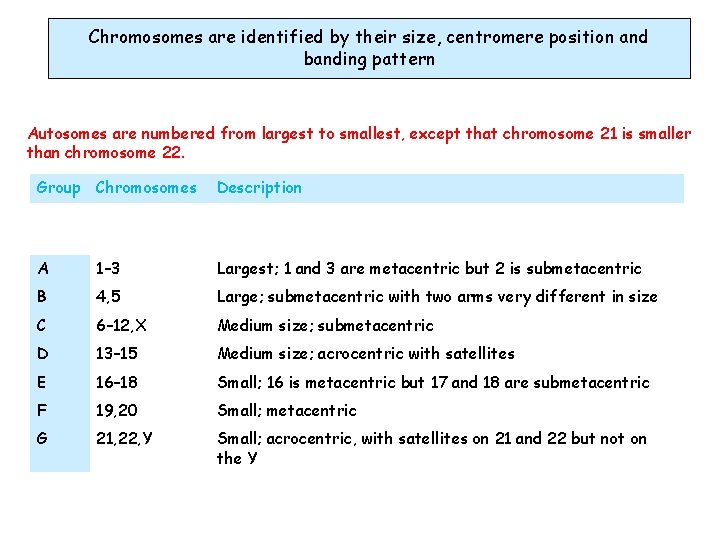
Chromosomes are identified by their size, centromere position and banding pattern Autosomes are numbered from largest to smallest, except that chromosome 21 is smaller than chromosome 22. Group Chromosomes Description A 1– 3 Largest; 1 and 3 are metacentric but 2 is submetacentric B 4, 5 Large; submetacentric with two arms very different in size C 6– 12, X Medium size; submetacentric D 13– 15 Medium size; acrocentric with satellites E 16– 18 Small; 16 is metacentric but 17 and 18 are submetacentric F 19, 20 Small; metacentric G 21, 22, Y Small; acrocentric, with satellites on 21 and 22 but not on the Y