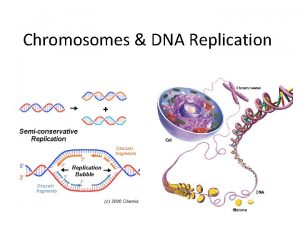
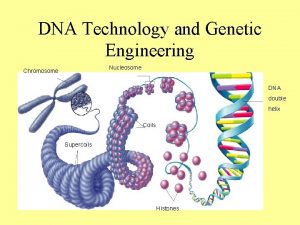
Chromosomes Chromatin and the Nucleosome Chromosomes DNA associated



- Slides: 49

Chromosomes, Chromatin, and the Nucleosome Chromosomes: DNA associated with proteins 1. The chromosome is a compact form of the DNA that readily fits inside the cell. 2. Packaging the DNA into chromosomes serves to protect the DNA from damage. 3. Only DNA packaged into a chromosome can be transmitted efficient to daughter cells.

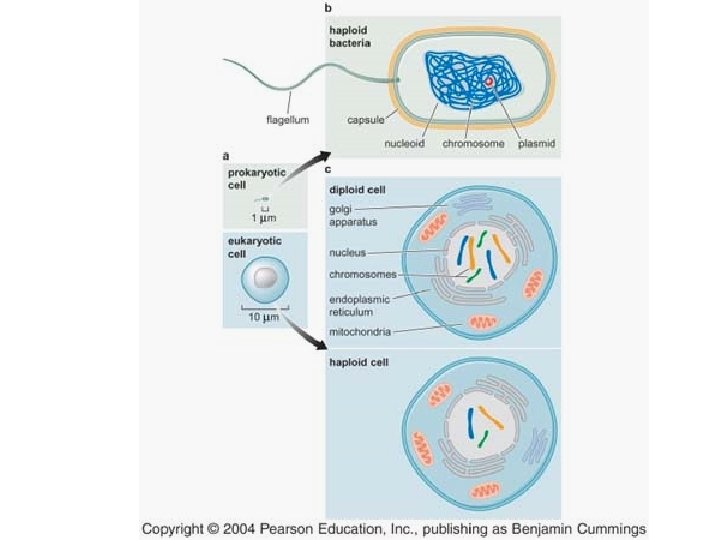
Table I: variation in chromosome makeup in different organisms The traditional view is that prokaryotic cells have a single, circular chromosome, and eukaryotic cells have multiple, linear chromosomes.


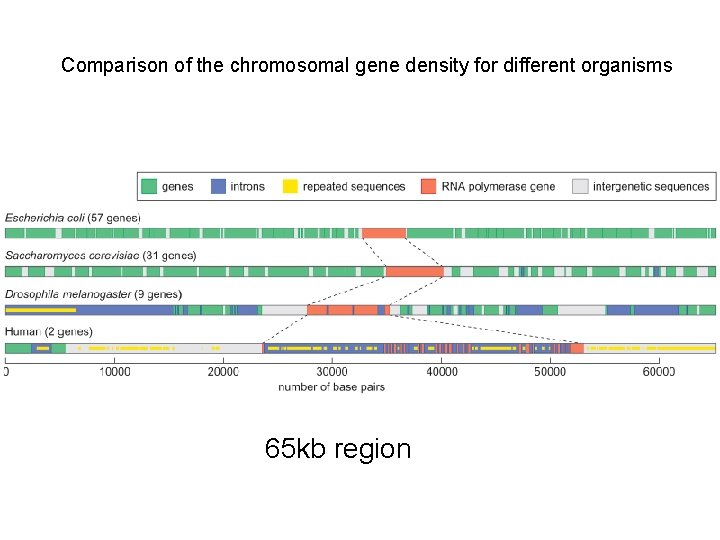
Table 2. Comparison of the gene density in different organisms’ genomes

Comparison of the chromosomal gene density for different organisms 65 kb region

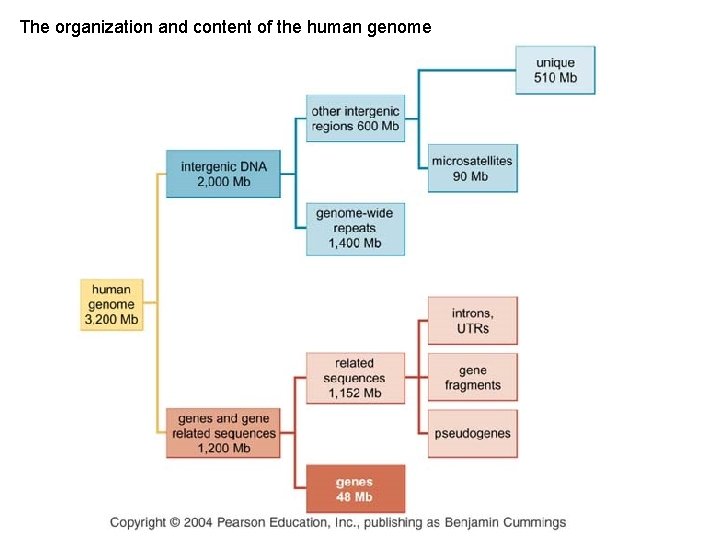
The organization and content of the human genome

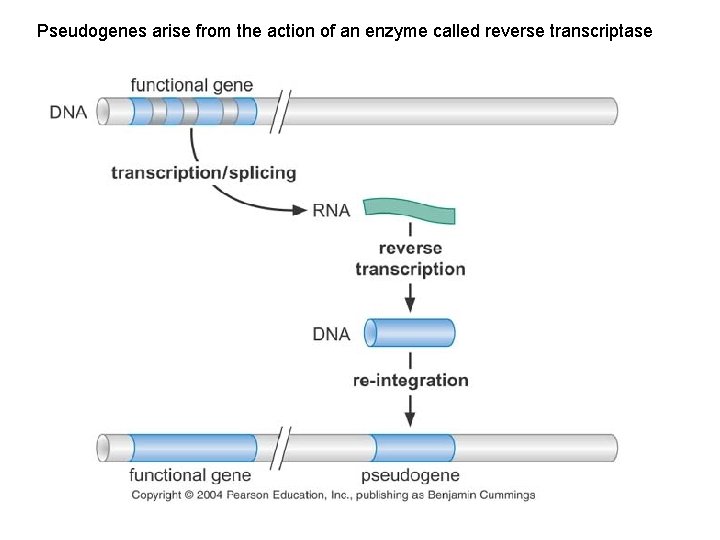
Pseudogenes arise from the action of an enzyme called reverse transcriptase

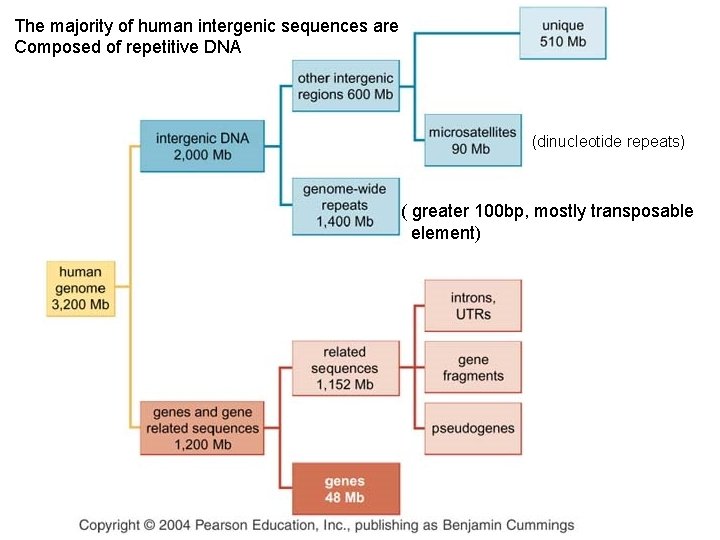
The majority of human intergenic sequences are Composed of repetitive DNA (dinucleotide repeats) ( greater 100 bp, mostly transposable element)

Table 7 -3 Contribution of introns and repeated sequences to different genomes introns (p. 135)

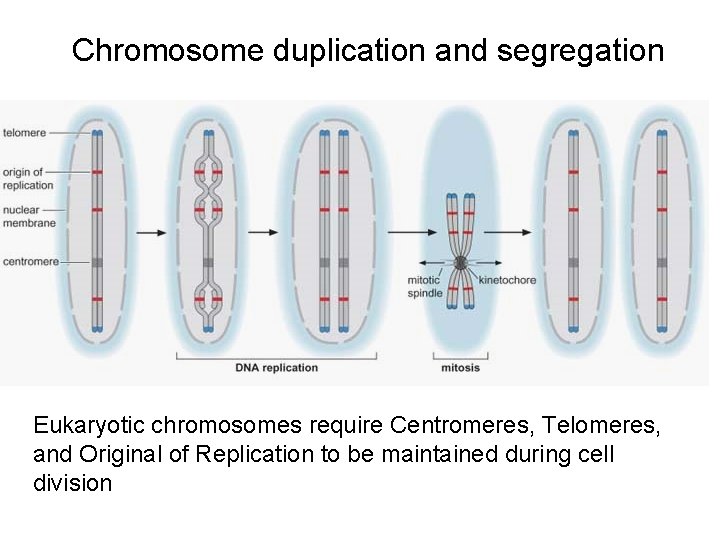
Chromosome duplication and segregation Eukaryotic chromosomes require Centromeres, Telomeres, and Original of Replication to be maintained during cell division

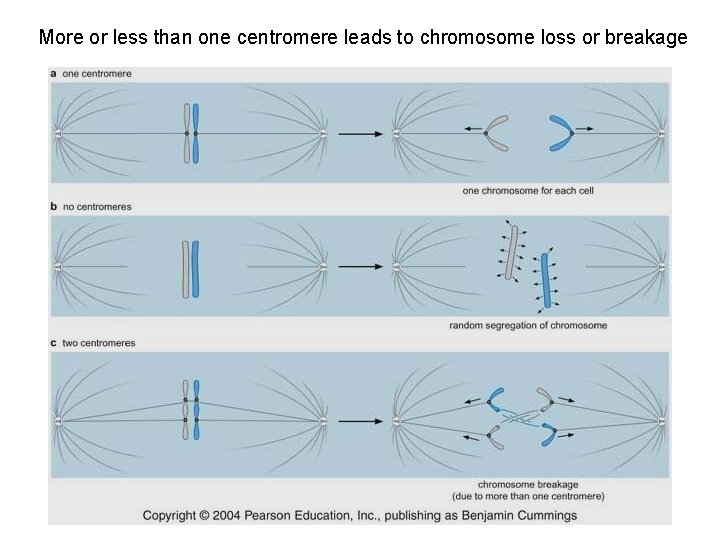
More or less than one centromere leads to chromosome loss or breakage

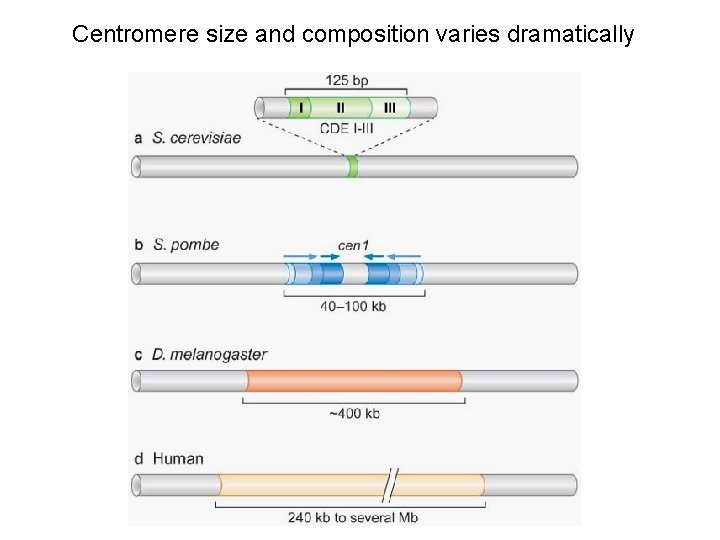
Centromere size and composition varies dramatically

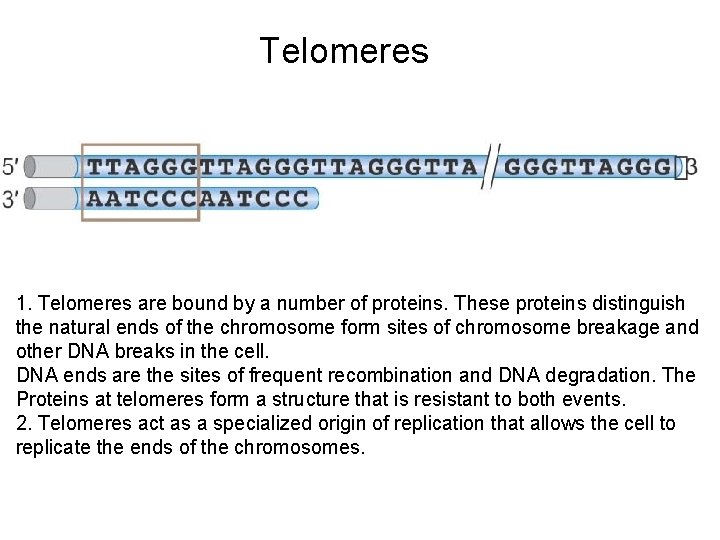
Telomeres 1. Telomeres are bound by a number of proteins. These proteins distinguish the natural ends of the chromosome form sites of chromosome breakage and other DNA breaks in the cell. DNA ends are the sites of frequent recombination and DNA degradation. The Proteins at telomeres form a structure that is resistant to both events. 2. Telomeres act as a specialized origin of replication that allows the cell to replicate the ends of the chromosomes.

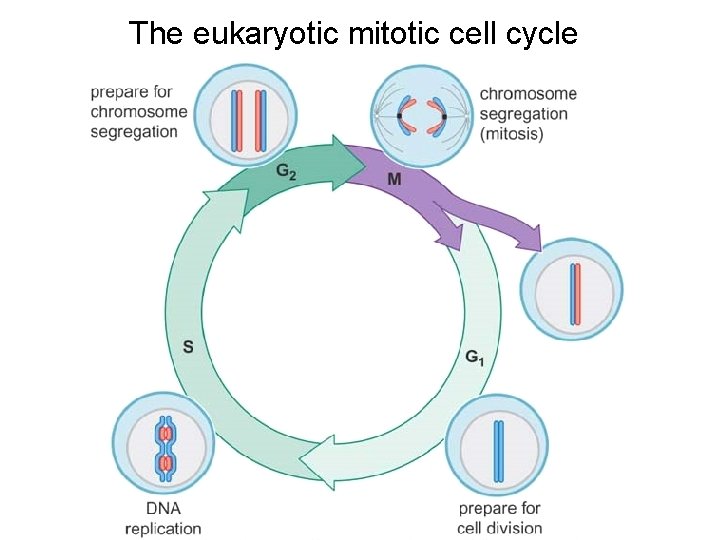
The eukaryotic mitotic cell cycle

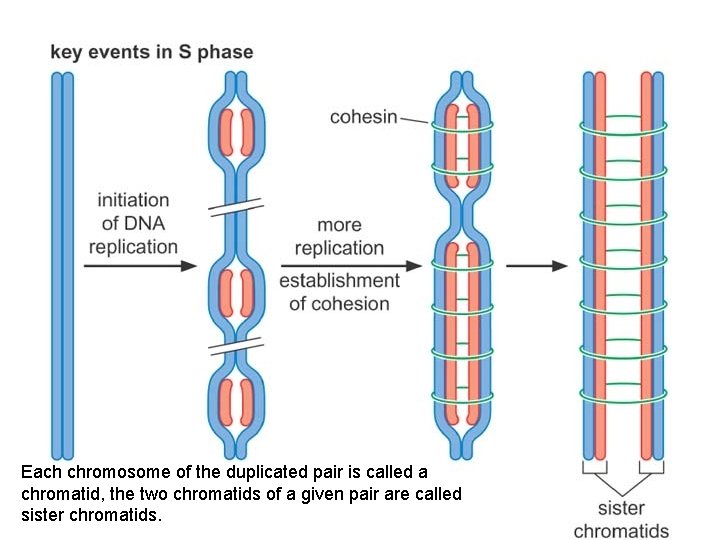
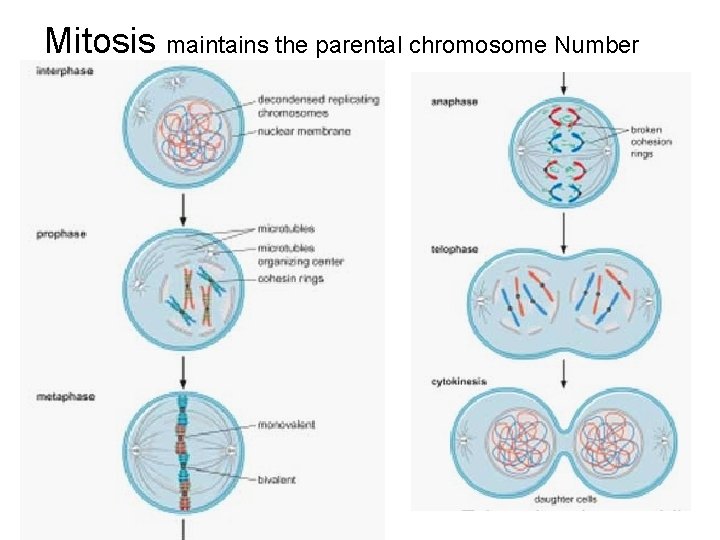
Each chromosome of the duplicated pair is called a chromatid, the two chromatids of a given pair are called sister chromatids.

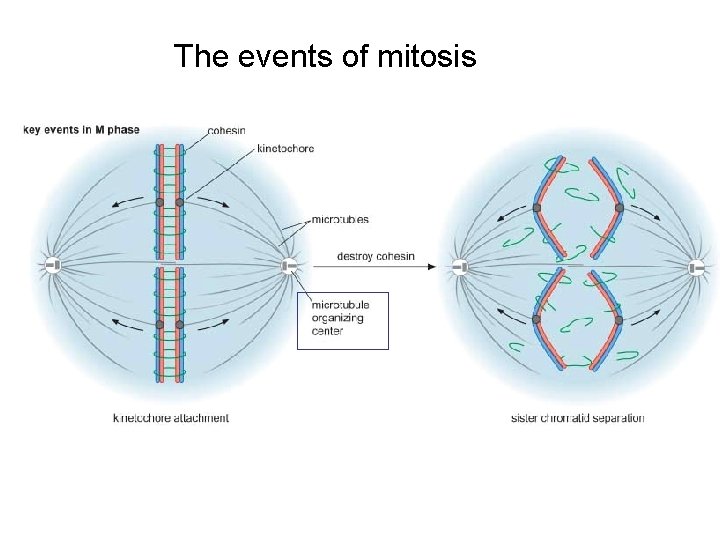
The events of mitosis

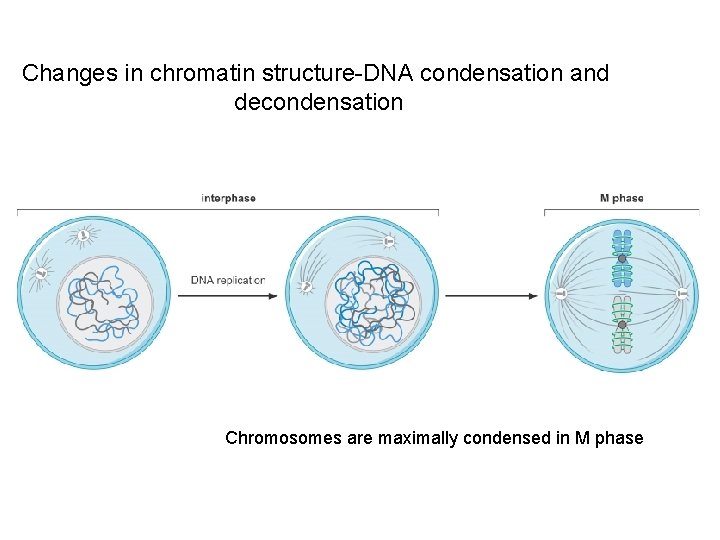
Changes in chromatin structure-DNA condensation and decondensation Chromosomes are maximally condensed in M phase

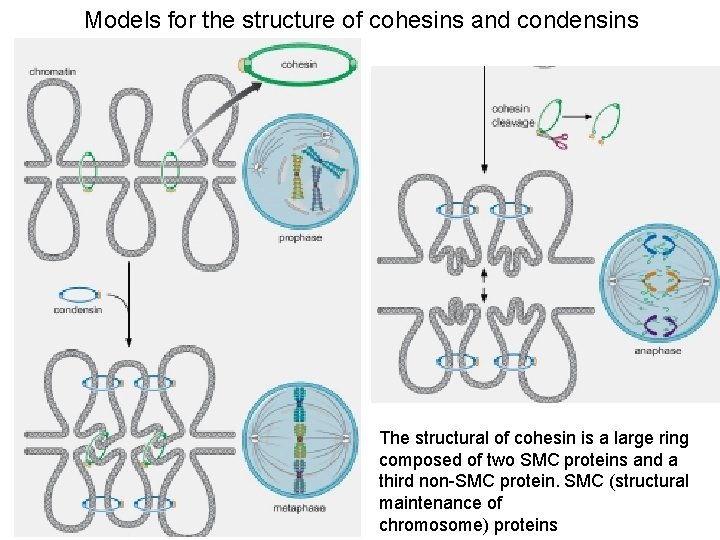
• Sister Chromatid cohension and Chromosome condensation are mediated by SMC ((structural maintenance of chromosome) proteins

Models for the structure of cohesins and condensins The structural of cohesin is a large ring composed of two SMC proteins and a third non-SMC protein. SMC (structural maintenance of chromosome) proteins

Mitosis maintains the parental chromosome Number

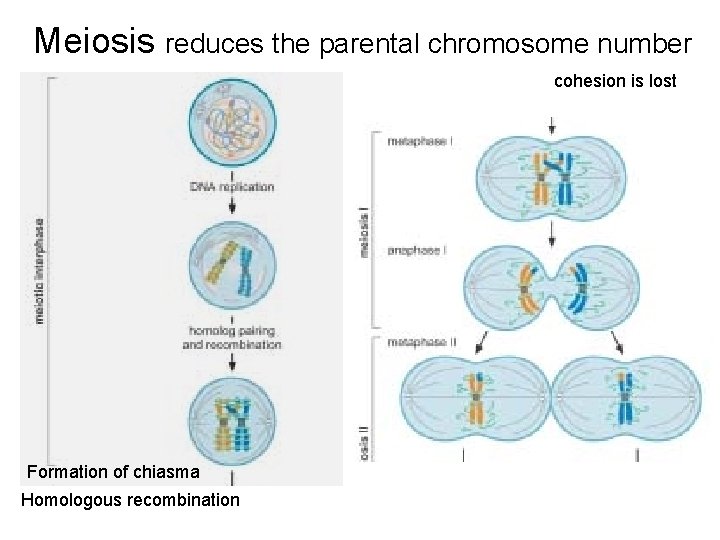
Meiosis reduces the parental chromosome number cohesion is lost Formation of chiasma Homologous recombination

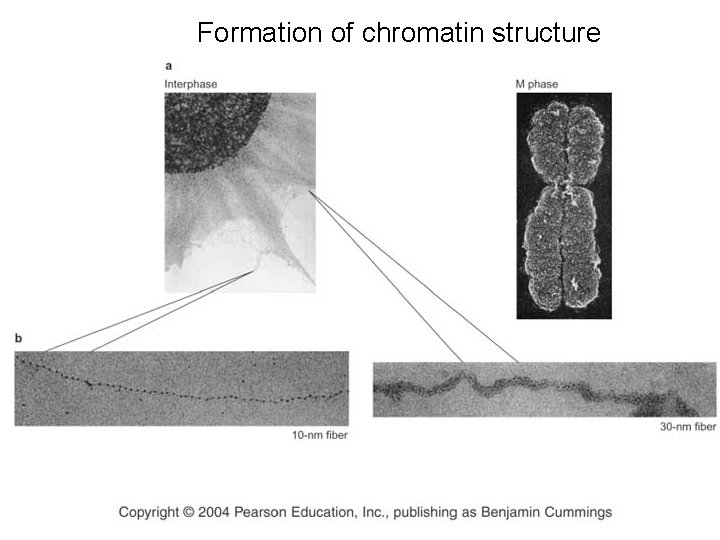
Formation of chromatin structure

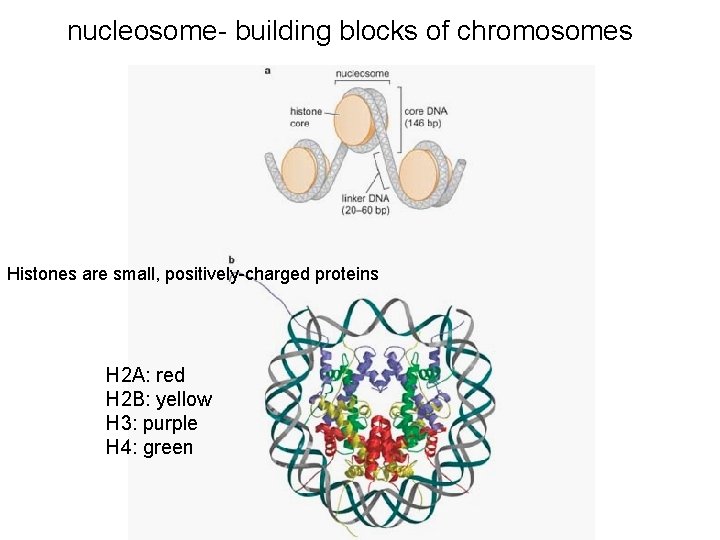
nucleosome- building blocks of chromosomes Histones are small, positively-charged proteins H 2 A: red H 2 B: yellow H 3: purple H 4: green


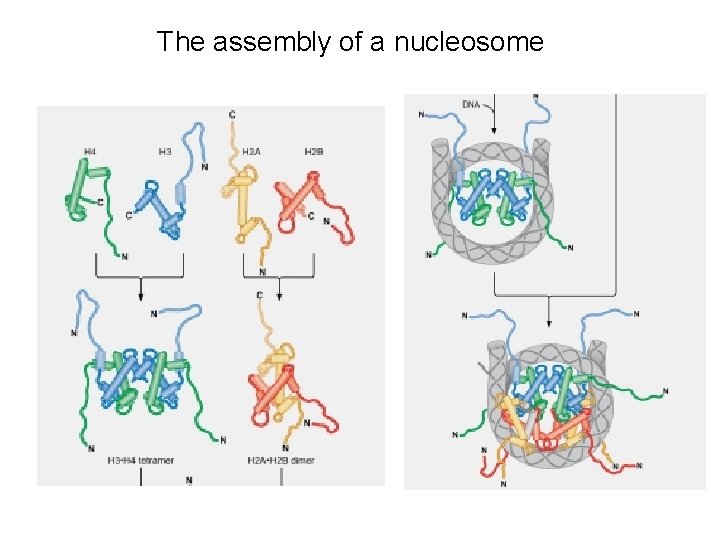
The assembly of a nucleosome

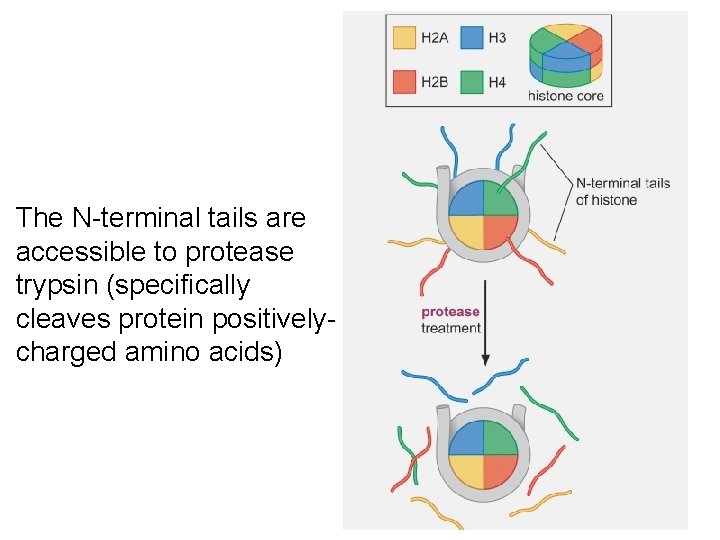
The N-terminal tails are accessible to protease trypsin (specifically cleaves protein positivelycharged amino acids)

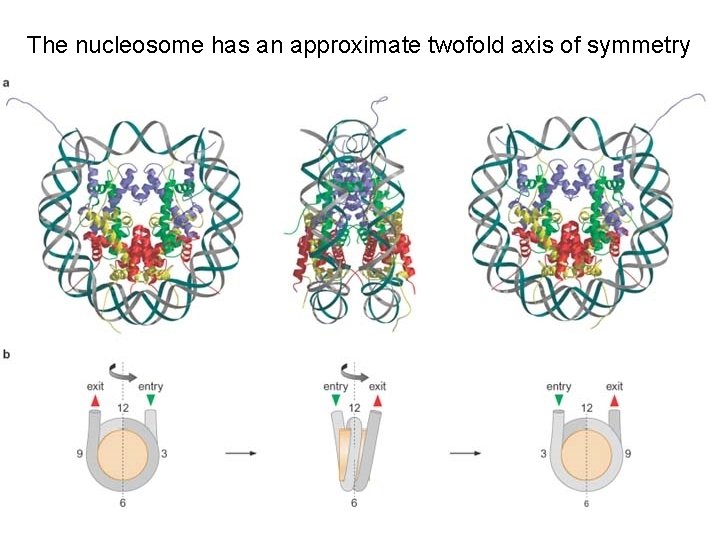
The nucleosome has an approximate twofold axis of symmetry

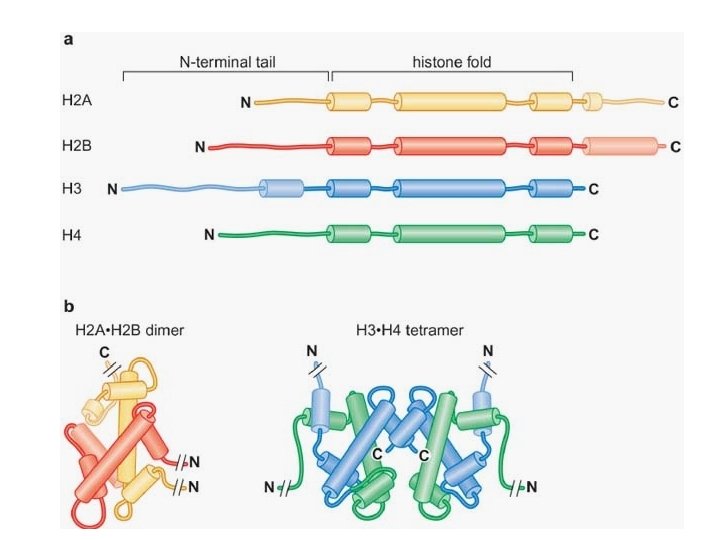
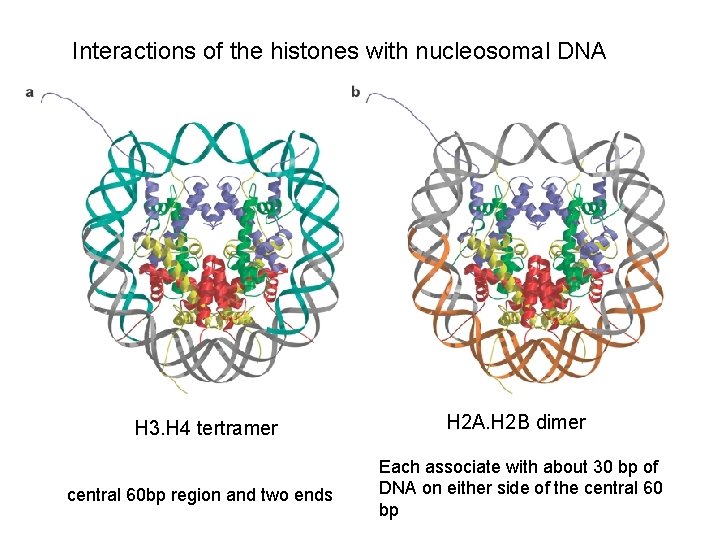
Interactions of the histones with nucleosomal DNA H 3. H 4 tertramer central 60 bp region and two ends H 2 A. H 2 B dimer Each associate with about 30 bp of DNA on either side of the central 60 bp

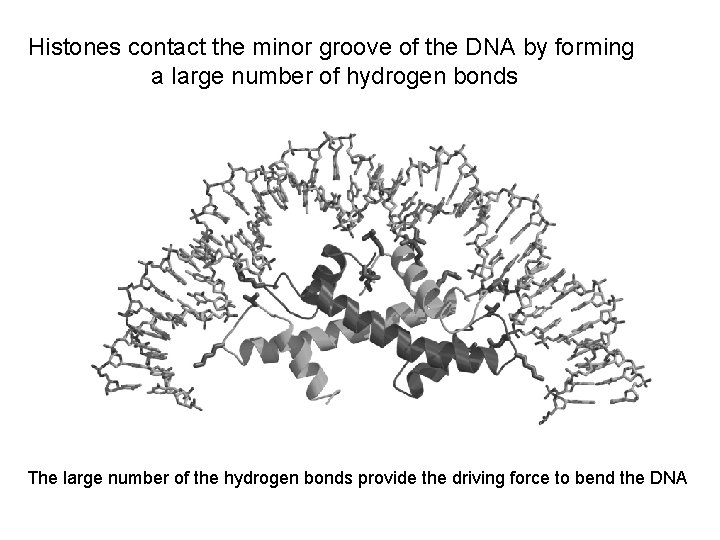
Histones contact the minor groove of the DNA by forming a large number of hydrogen bonds The large number of the hydrogen bonds provide the driving force to bend the DNA

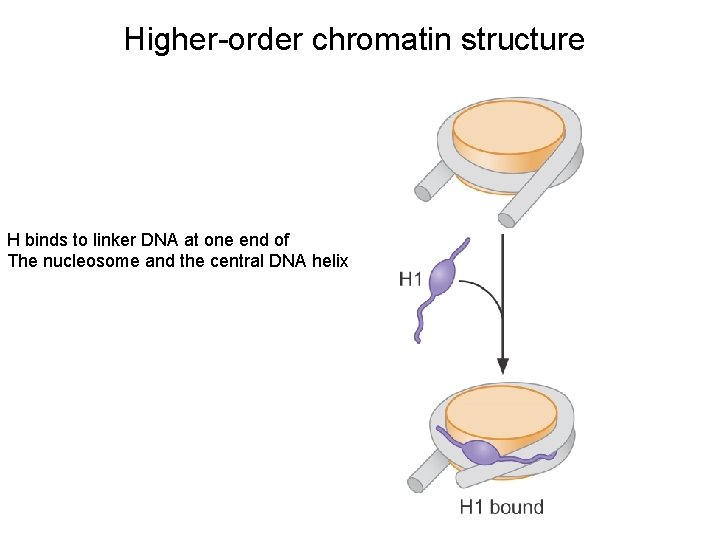
Higher-order chromatin structure H binds to linker DNA at one end of The nucleosome and the central DNA helix

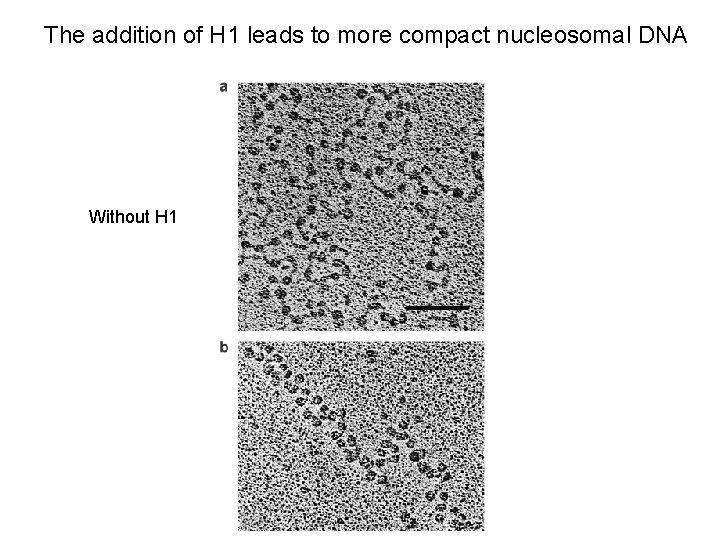
The addition of H 1 leads to more compact nucleosomal DNA Without H 1

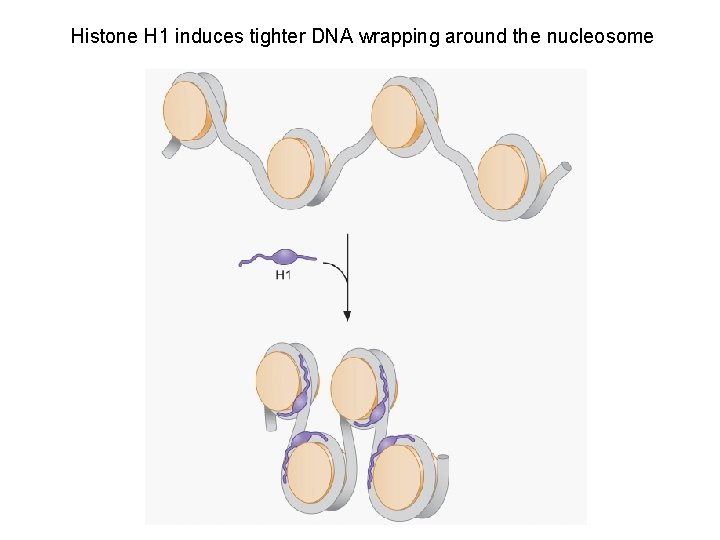
Histone H 1 induces tighter DNA wrapping around the nucleosome

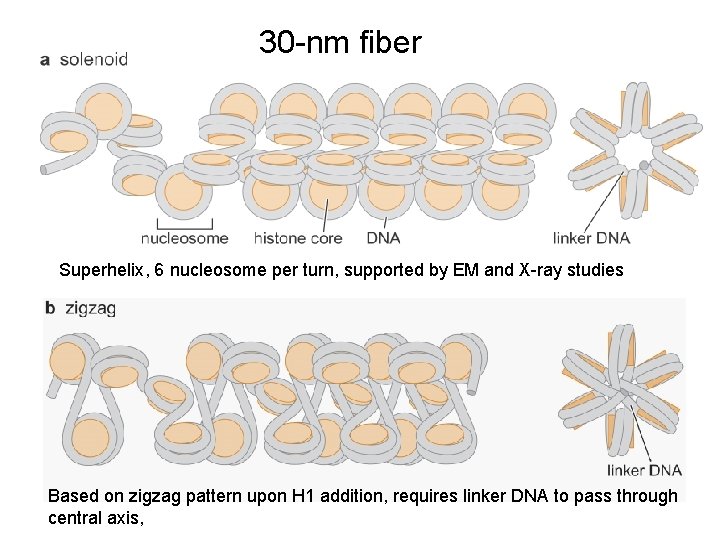
30 -nm fiber Superhelix, 6 nucleosome per turn, supported by EM and X-ray studies Based on zigzag pattern upon H 1 addition, requires linker DNA to pass through central axis,

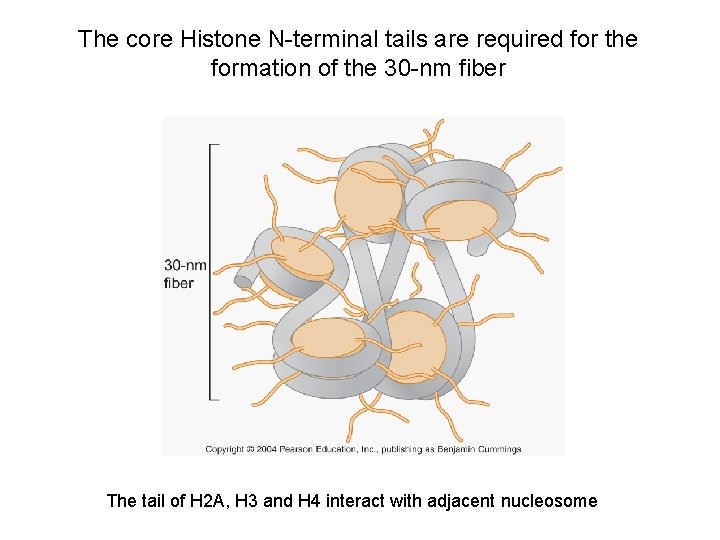
The core Histone N-terminal tails are required for the formation of the 30 -nm fiber The tail of H 2 A, H 3 and H 4 interact with adjacent nucleosome

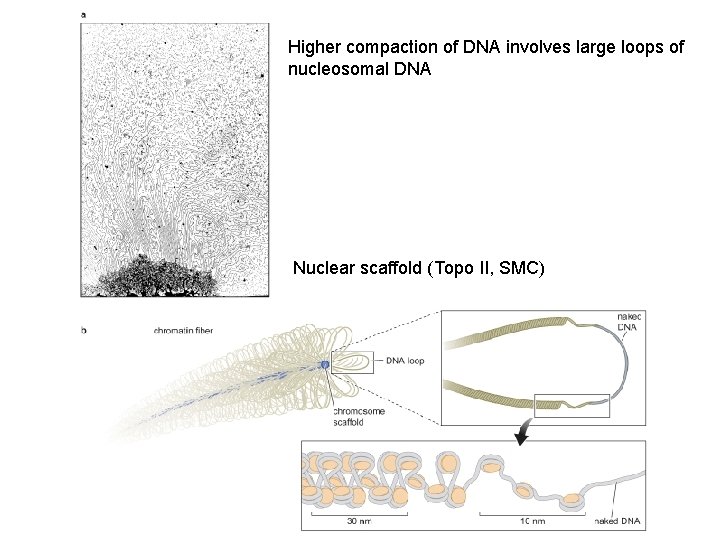
Higher compaction of DNA involves large loops of nucleosomal DNA Nuclear scaffold (Topo II, SMC)

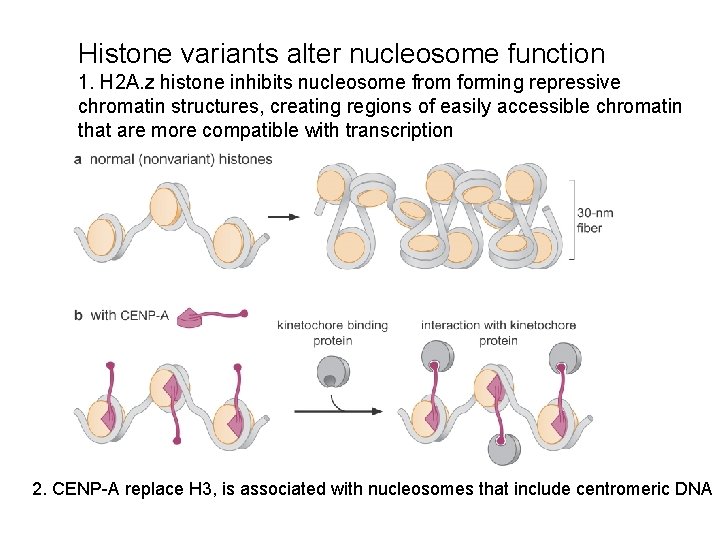
Histone variants alter nucleosome function 1. H 2 A. z histone inhibits nucleosome from forming repressive chromatin structures, creating regions of easily accessible chromatin that are more compatible with transcription 2. CENP-A replace H 3, is associated with nucleosomes that include centromeric DNA

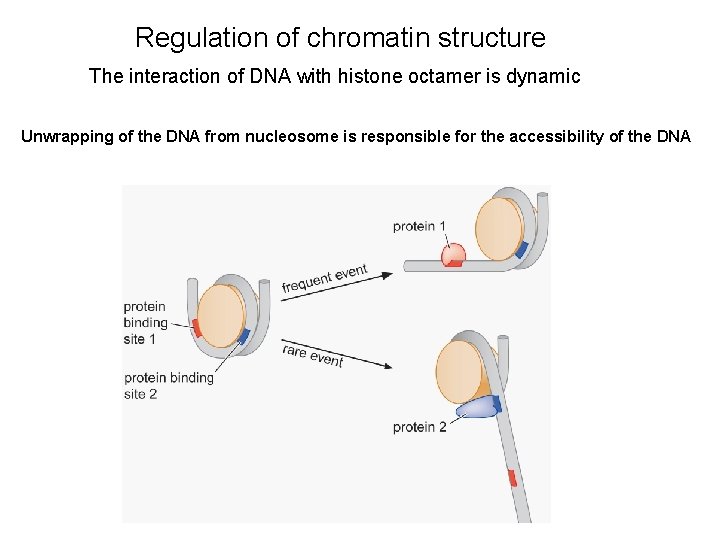
Regulation of chromatin structure The interaction of DNA with histone octamer is dynamic Unwrapping of the DNA from nucleosome is responsible for the accessibility of the DNA

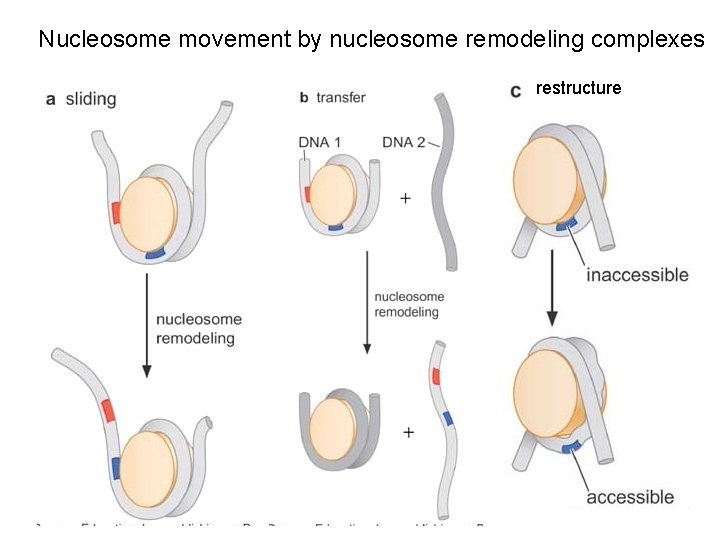
Nucleosome movement by nucleosome remodeling complexes restructure

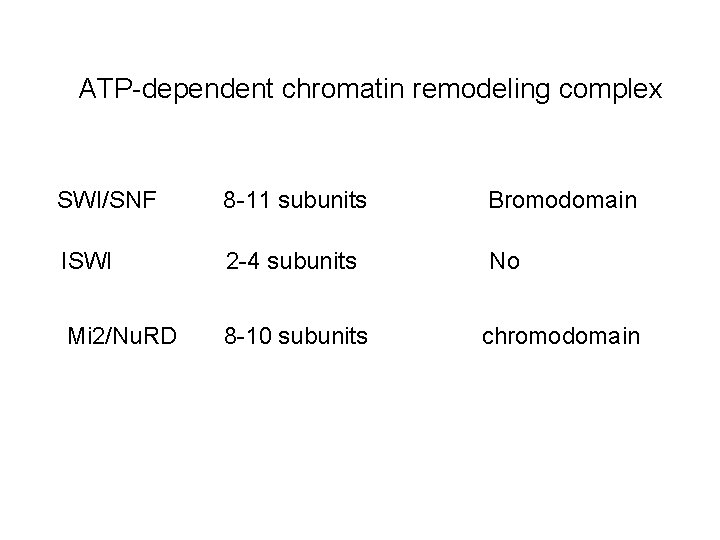
ATP-dependent chromatin remodeling complex SWI/SNF 8 -11 subunits Bromodomain ISWI 2 -4 subunits No Mi 2/Nu. RD 8 -10 subunits chromodomain

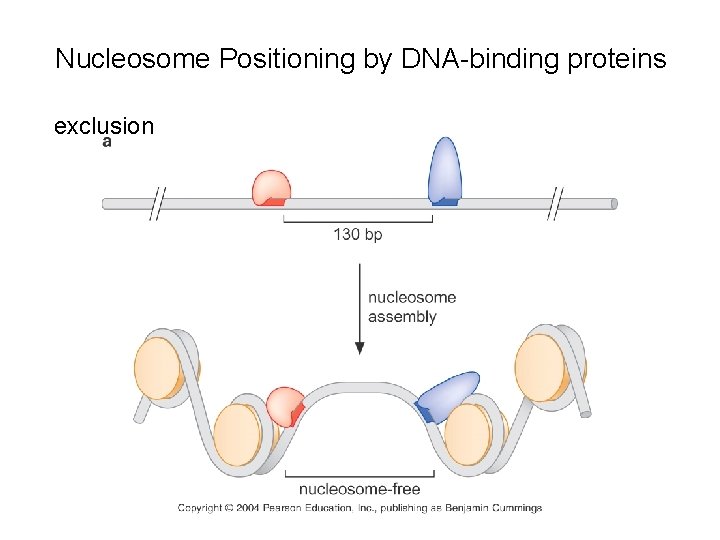
Nucleosome Positioning by DNA-binding proteins exclusion

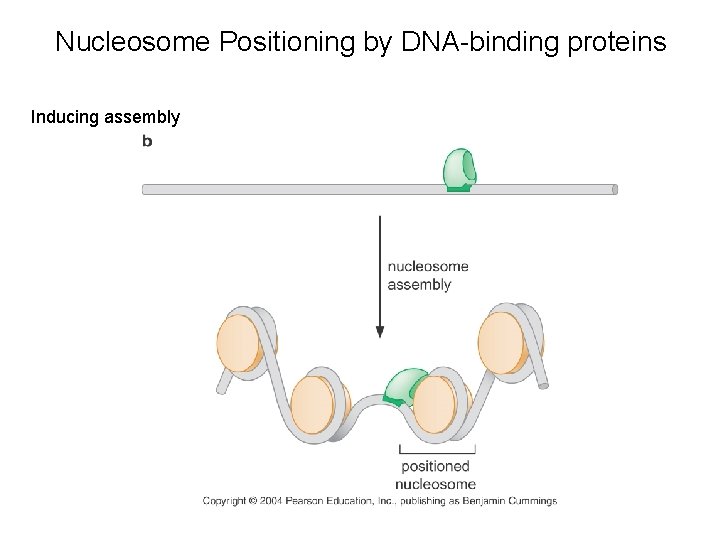
Nucleosome Positioning by DNA-binding proteins Inducing assembly

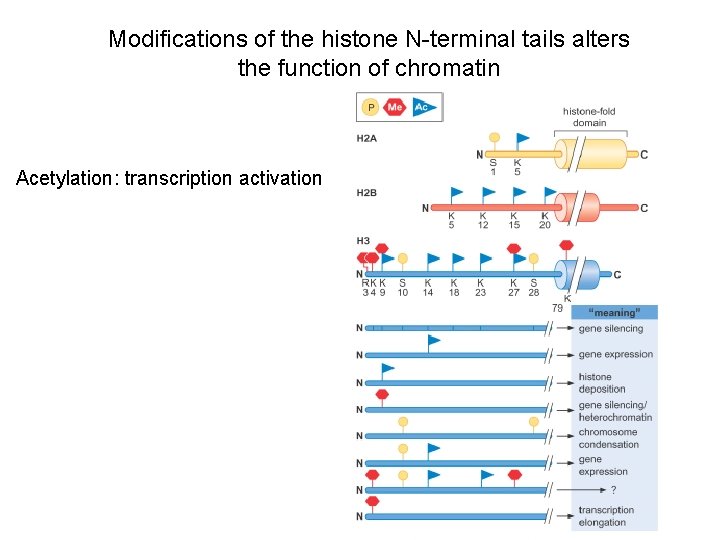
Modifications of the histone N-terminal tails alters the function of chromatin Acetylation: transcription activation

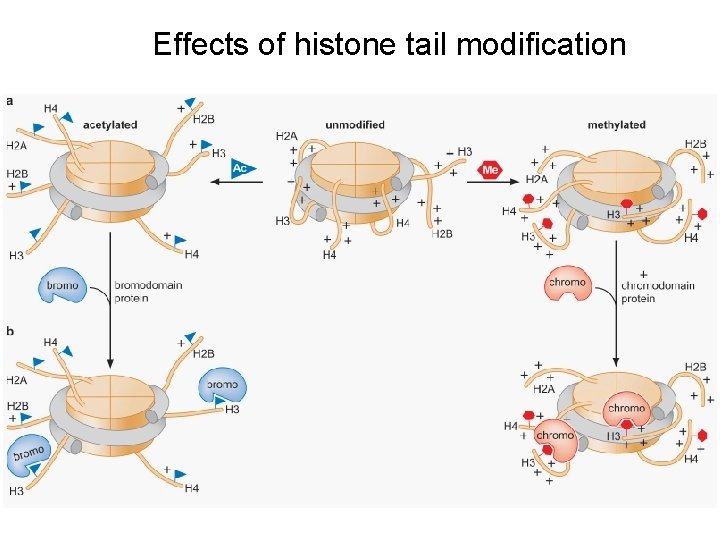
Effects of histone tail modification

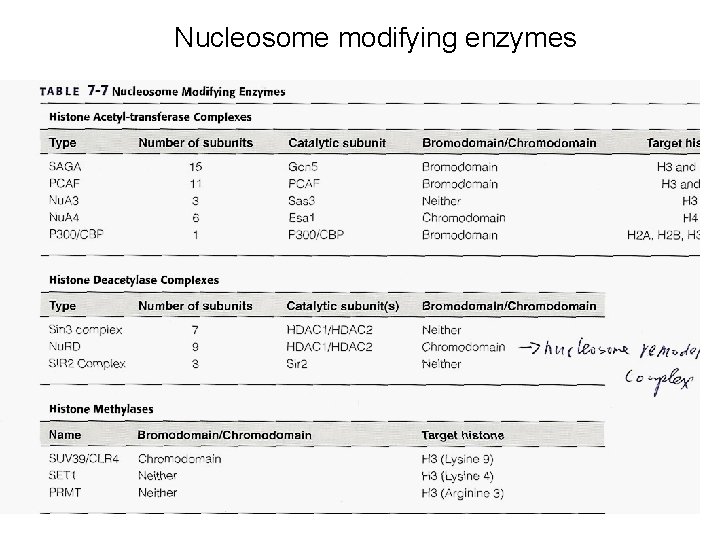
Nucleosome modifying enzymes

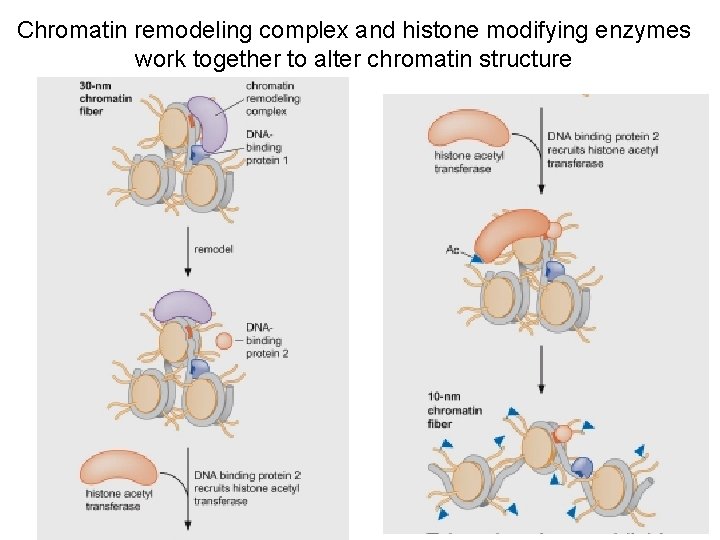
Chromatin remodeling complex and histone modifying enzymes work together to alter chromatin structure

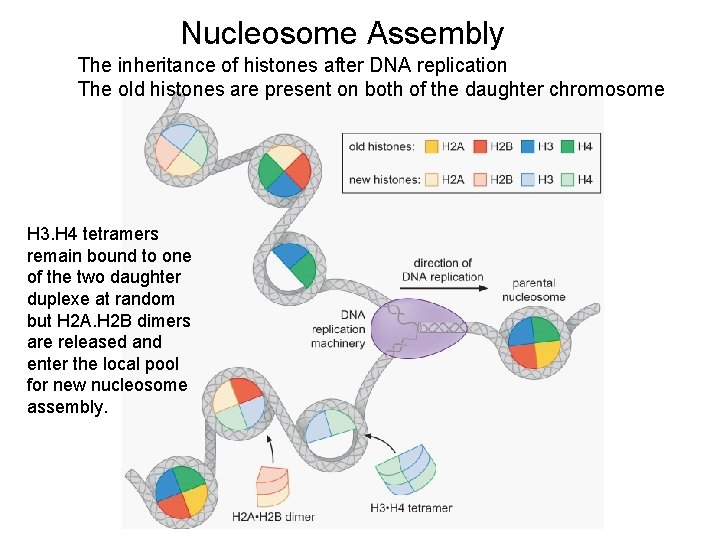
Nucleosome Assembly The inheritance of histones after DNA replication The old histones are present on both of the daughter chromosome H 3. H 4 tetramers remain bound to one of the two daughter duplexe at random but H 2 A. H 2 B dimers are released and enter the local pool for new nucleosome assembly.

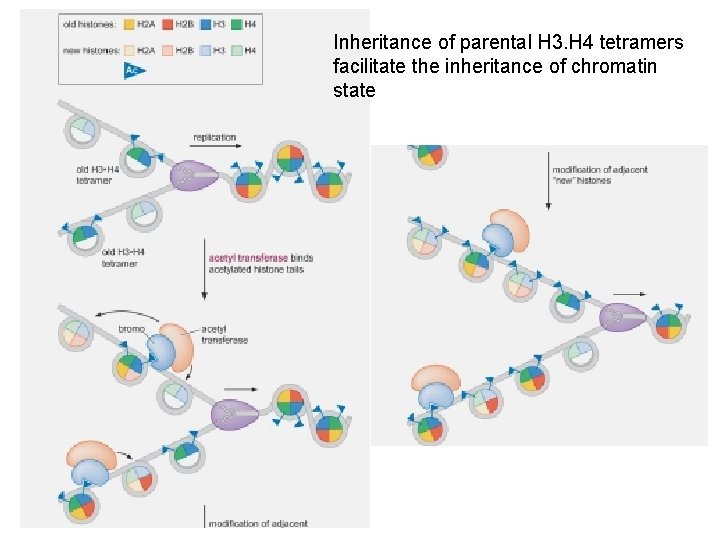
Inheritance of parental H 3. H 4 tetramers facilitate the inheritance of chromatin state

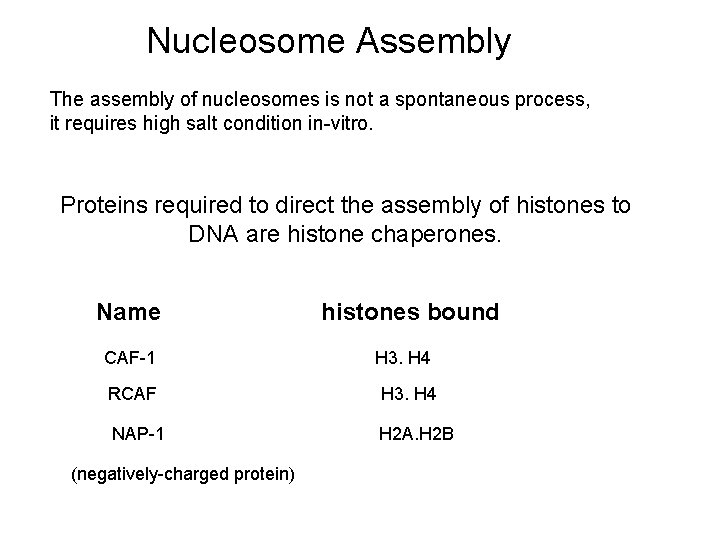
Nucleosome Assembly The assembly of nucleosomes is not a spontaneous process, it requires high salt condition in-vitro. Proteins required to direct the assembly of histones to DNA are histone chaperones. Name CAF-1 histones bound H 3. H 4 RCAF H 3. H 4 NAP-1 H 2 A. H 2 B (negatively-charged protein)

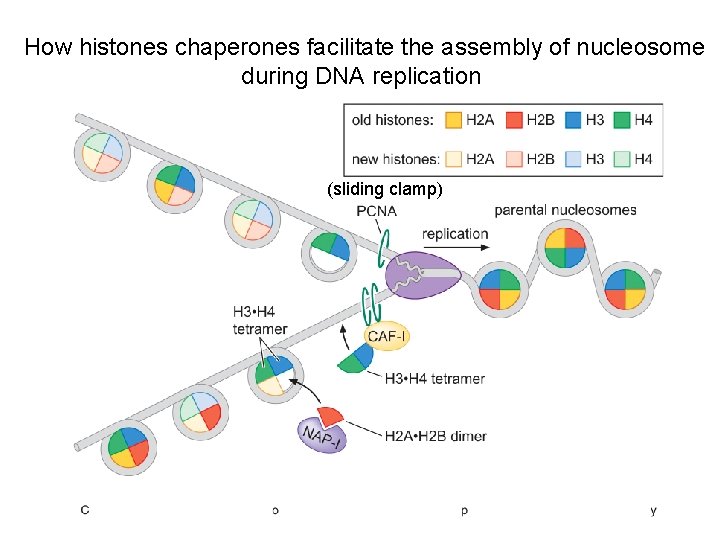
How histones chaperones facilitate the assembly of nucleosome during DNA replication (sliding clamp)
 Nucleosome remodeling
Nucleosome remodeling Nucleosome model
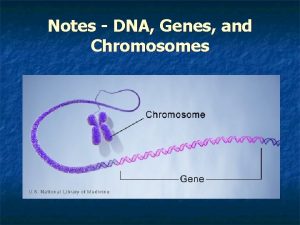
Nucleosome model Genes chromosomes and dna
Genes chromosomes and dna Building vocabulary: the nucleus, dna, and chromosomes
Building vocabulary: the nucleus, dna, and chromosomes Dna, genes and chromosomes relationship
Dna, genes and chromosomes relationship Dna scrunches up and chromosomes are first visible
Dna scrunches up and chromosomes are first visible What is the relationship between dna chromosomes and genes
What is the relationship between dna chromosomes and genes Organism
Organism Chromatin vs chromozom
Chromatin vs chromozom Chromatin vs chromozom
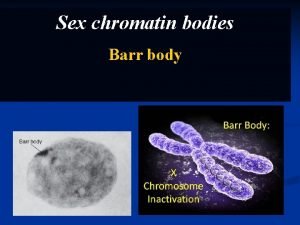
Chromatin vs chromozom Barrovo telisko
Barrovo telisko Chromatin body
Chromatin body Chromatin in a sentence
Chromatin in a sentence Dot
Dot Primodial germ cell
Primodial germ cell Parasites
Parasites Chromatin draws together to create
Chromatin draws together to create Chromatin
Chromatin Chromatin
Chromatin Chromatin states
Chromatin states Chromatin immunoprecipitation
Chromatin immunoprecipitation Chromatin packing
Chromatin packing Coding dna and non coding dna
Coding dna and non coding dna Function of chromosome
Function of chromosome Genes chromosome
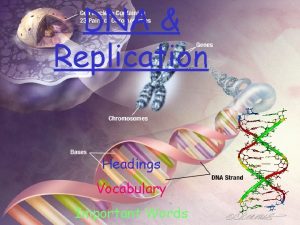
Genes chromosome Dna chromosomes genes diagram
Dna chromosomes genes diagram Replication fork
Replication fork Bioflix activity dna replication nucleotide pairing
Bioflix activity dna replication nucleotide pairing What role does dna polymerase play in copying dna?
What role does dna polymerase play in copying dna? Chapter 11 dna and genes
Chapter 11 dna and genes Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Frameset trong html5
Frameset trong html5 Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Chó sói
Chó sói Tư thế worms-breton
Tư thế worms-breton Chúa yêu trần thế alleluia
Chúa yêu trần thế alleluia Các môn thể thao bắt đầu bằng tiếng bóng
Các môn thể thao bắt đầu bằng tiếng bóng Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Cong thức tính động năng
Cong thức tính động năng Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Mật thư anh em như thể tay chân
Mật thư anh em như thể tay chân 101012 bằng
101012 bằng Phản ứng thế ankan
Phản ứng thế ankan Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Thơ thất ngôn tứ tuyệt đường luật
Thơ thất ngôn tứ tuyệt đường luật Quá trình desamine hóa có thể tạo ra
Quá trình desamine hóa có thể tạo ra Một số thể thơ truyền thống
Một số thể thơ truyền thống Cái miệng nó xinh thế chỉ nói điều hay thôi
Cái miệng nó xinh thế chỉ nói điều hay thôi