Chromatin state dynamics in nine human cell types


- Slides: 18

Chromatin state dynamics in nine human cell types elucidate regulators and disease-associated SNPs Jason Ernst Joint work with Pouya Kheradpour, Luke Ward Brad Bernstein and Manolis Kellis

Challenge: interpreting disease-associated variants CATGACTG CATGCCTG Epigenomics Disease variants • GWAS studies implicate thousands of non-coding loci associated with disease • Challenges towards interpreting disease variants: – Find ‘true’ causative SNP among many candidates in LD – Determining type of function: especially outside protein-coding – Reveal relevant cell type of activity – Link to upstream regulators and downstream targets • This talk: chromatin tools to address these challenges 2

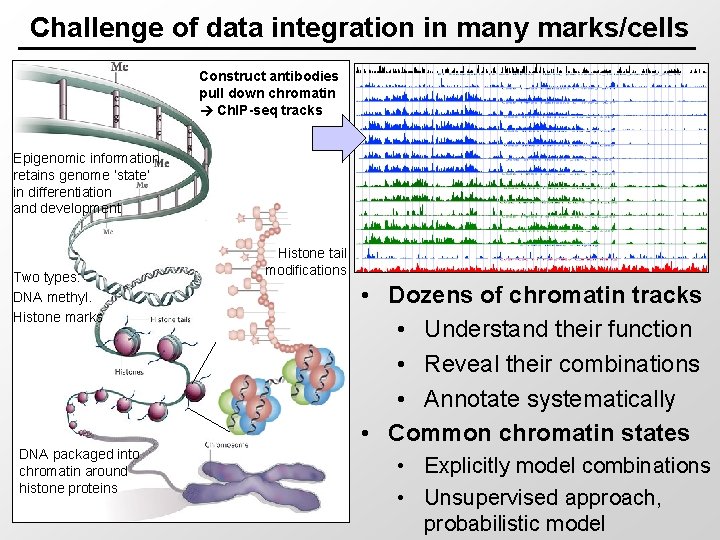
Challenge of data integration in many marks/cells Construct antibodies pull down chromatin Ch. IP-seq tracks Epigenomic information retains genome ‘state’ in differentiation and development Two types: DNA methyl. Histone marks DNA packaged into chromatin around histone proteins Histone tail modifications • Dozens of chromatin tracks • Understand their function • Reveal their combinations • Annotate systematically • Common chromatin states • Explicitly model combinations • Unsupervised approach, probabilistic model

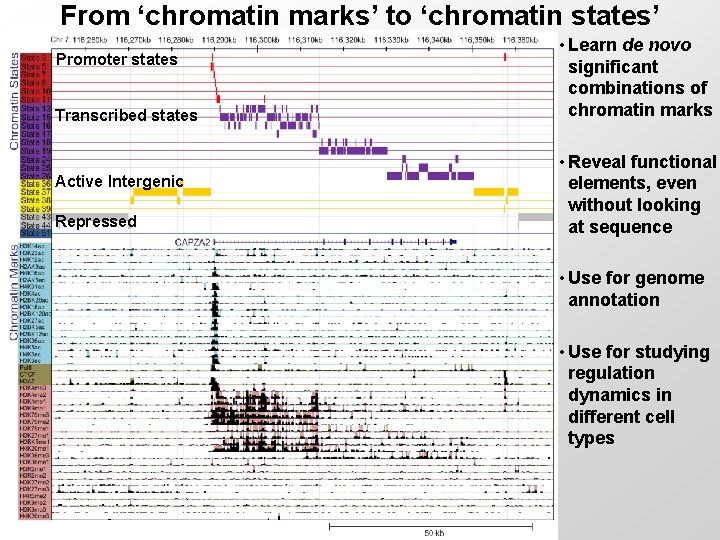
From ‘chromatin marks’ to ‘chromatin states’ Promoter states Transcribed states Active Intergenic Repressed • Learn de novo significant combinations of chromatin marks • Reveal functional elements, even without looking at sequence • Use for genome annotation • Use for studying regulation dynamics in different cell types

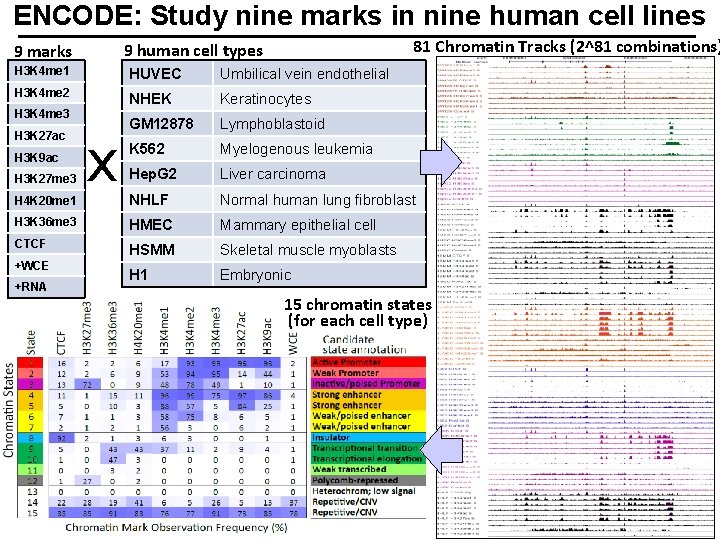
ENCODE: Study nine marks in nine human cell lines 81 Chromatin Tracks (2^81 combinations) 9 human cell types 9 marks H 3 K 4 me 1 HUVEC Umbilical vein endothelial H 3 K 4 me 2 NHEK Keratinocytes GM 12878 Lymphoblastoid K 562 Myelogenous leukemia Hep. G 2 Liver carcinoma NHLF Normal human lung fibroblast H 3 K 36 me 3 HMEC Mammary epithelial cell CTCF HSMM Skeletal muscle myoblasts H 1 Embryonic H 3 K 4 me 3 H 3 K 27 ac H 3 K 9 ac H 3 K 27 me 3 H 4 K 20 me 1 +WCE +RNA x 15 chromatin states (for each cell type)

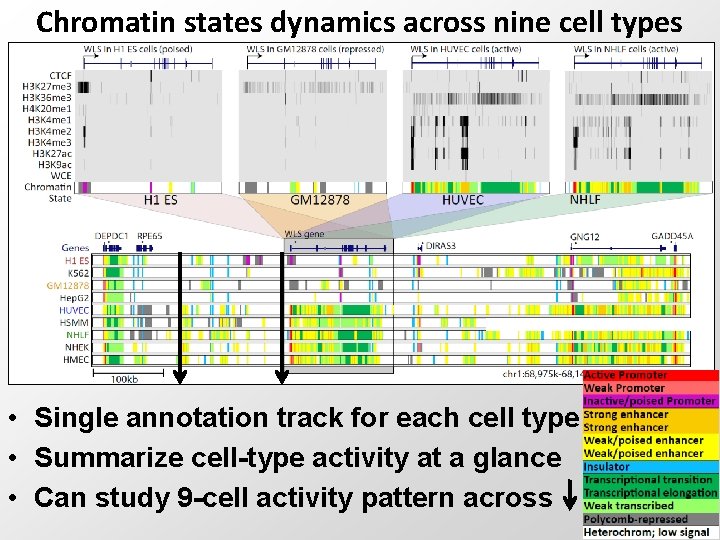
Chromatin states dynamics across nine cell types • Single annotation track for each cell type • Summarize cell-type activity at a glance • Can study 9 -cell activity pattern across

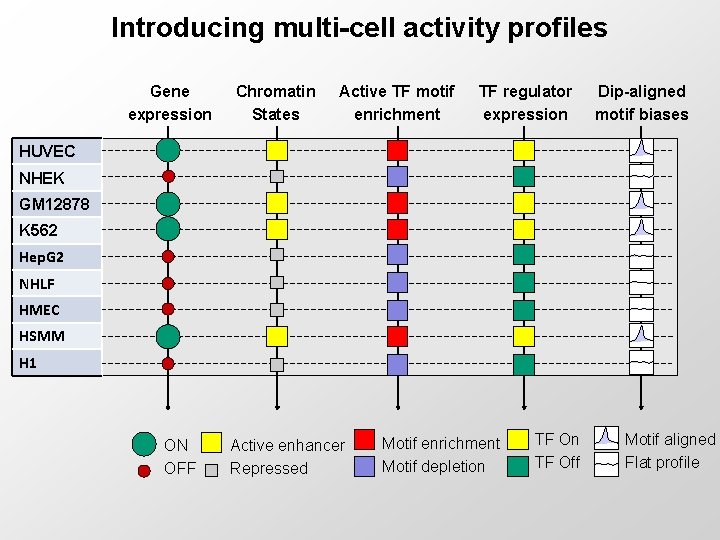
Introducing multi-cell activity profiles Gene expression Chromatin States Active TF motif enrichment TF regulator expression Dip-aligned motif biases HUVEC NHEK GM 12878 K 562 Hep. G 2 NHLF HMEC HSMM H 1 ON OFF Active enhancer Repressed Motif enrichment Motif depletion TF Off Motif aligned Flat profile

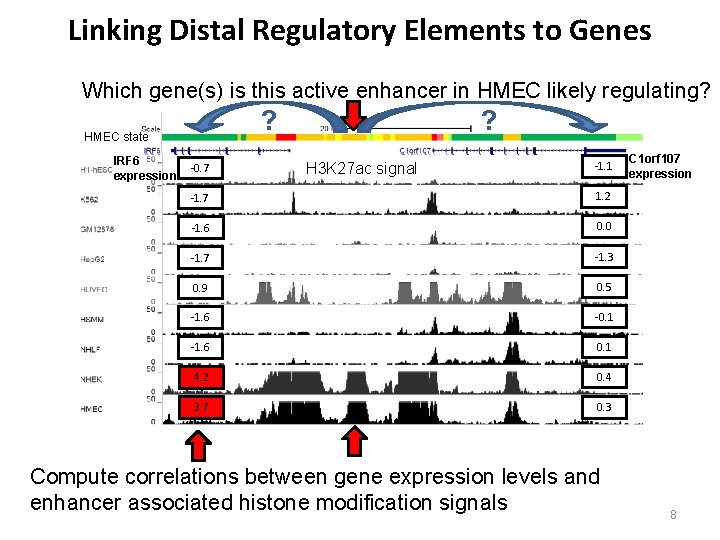
Linking Distal Regulatory Elements to Genes Which gene(s) is this active enhancer in HMEC likely regulating? ? HMEC state IRF 6 expression -0. 7 ? H 3 K 27 ac signal -1. 1 -1. 7 1. 2 -1. 6 0. 0 -1. 7 -1. 3 0. 9 0. 5 -1. 6 -0. 1 -1. 6 0. 1 4. 2 0. 4 3. 7 0. 3 Compute correlations between gene expression levels and enhancer associated histone modification signals C 1 orf 107 expression 8

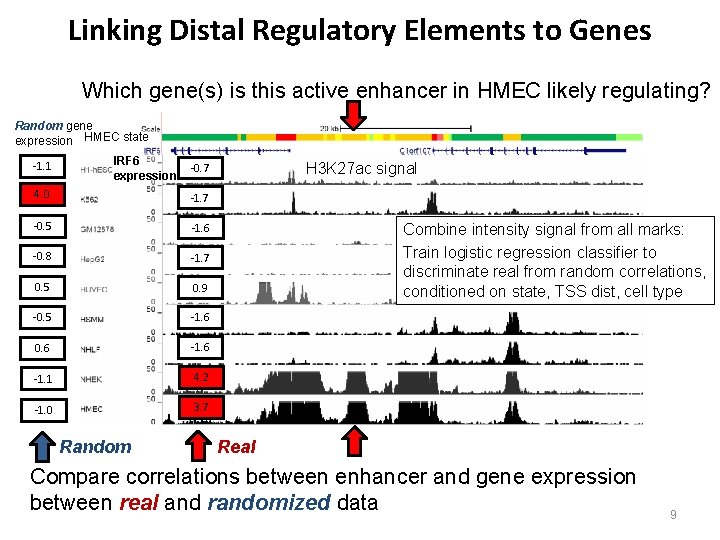
Linking Distal Regulatory Elements to Genes Which gene(s) is this active enhancer in HMEC likely regulating? Random gene expression HMEC state -1. 1 IRF 6 expression 4. 0 -1. 7 -0. 5 -1. 6 -0. 8 -1. 7 0. 5 0. 9 -0. 5 -1. 6 0. 6 -1. 1 4. 2 -1. 0 3. 7 Random H 3 K 27 ac signal -0. 7 Combine intensity signal from all marks: Train logistic regression classifier to discriminate real from random correlations, conditioned on state, TSS dist, cell type Real Compare correlations between enhancer and gene expression between real and randomized data 9

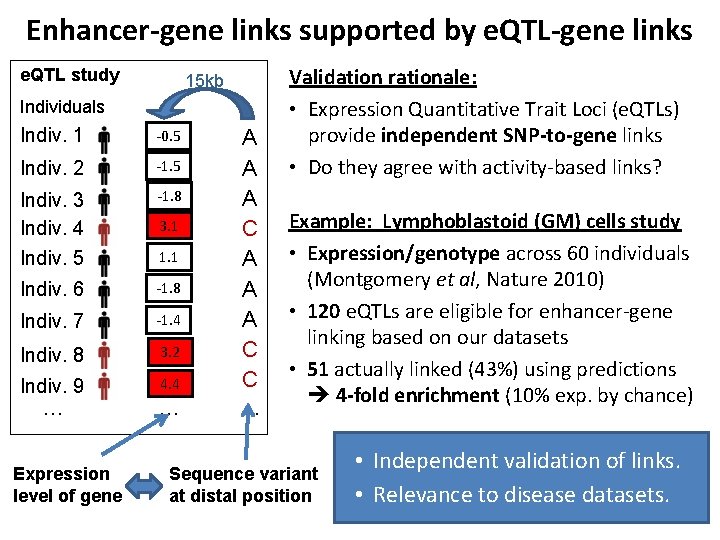
Enhancer-gene links supported by e. QTL-gene links e. QTL study 15 kb Individuals Indiv. 1 -0. 5 Indiv. 2 -1. 5 Indiv. 3 Indiv. 4 -1. 8 Indiv. 5 1. 1 Indiv. 6 -1. 8 Indiv. 7 -1. 4 Indiv. 8 3. 2 Indiv. 9 4. 4 … Expression level of gene 3. 1 … A A A C C … Validation rationale: • Expression Quantitative Trait Loci (e. QTLs) provide independent SNP-to-gene links • Do they agree with activity-based links? Example: Lymphoblastoid (GM) cells study • Expression/genotype across 60 individuals (Montgomery et al, Nature 2010) • 120 e. QTLs are eligible for enhancer-gene linking based on our datasets • 51 actually linked (43%) using predictions 4 -fold enrichment (10% exp. by chance) Sequence variant at distal position • Independent validation of links. • Relevance to disease datasets. 10

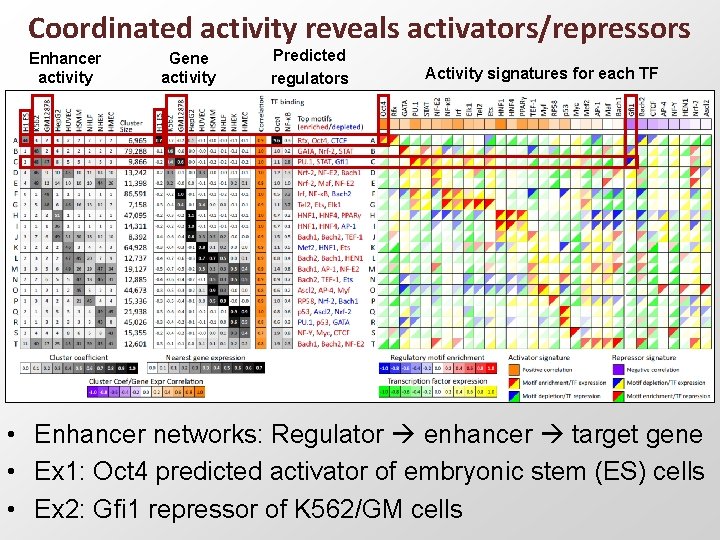
Coordinated activity reveals activators/repressors Enhancer activity Gene activity Predicted regulators Activity signatures for each TF • Enhancer networks: Regulator enhancer target gene • Ex 1: Oct 4 predicted activator of embryonic stem (ES) cells • Ex 2: Gfi 1 repressor of K 562/GM cells

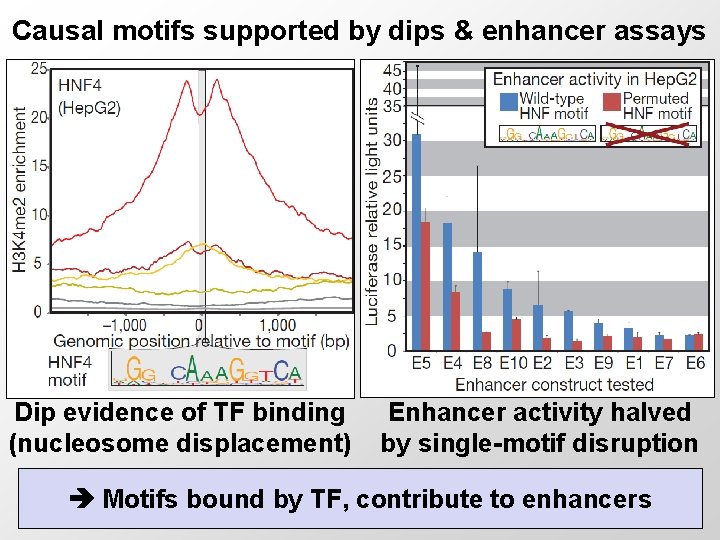
Causal motifs supported by dips & enhancer assays Dip evidence of TF binding (nucleosome displacement) Enhancer activity halved by single-motif disruption Motifs bound by TF, contribute to enhancers 12

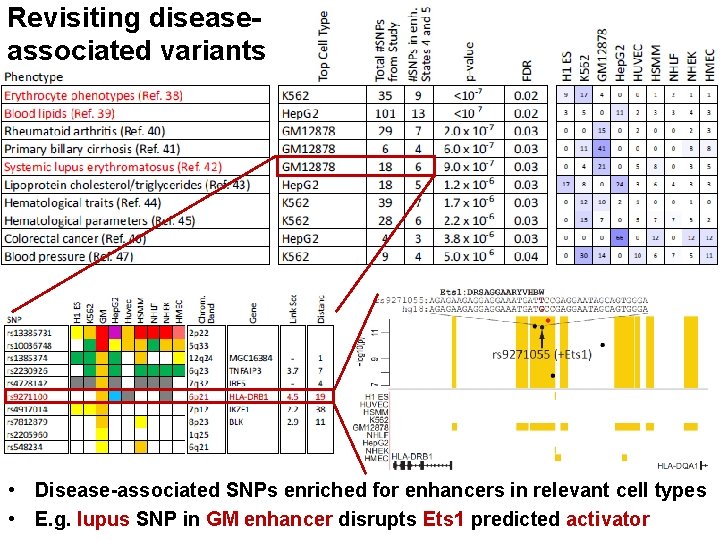
Revisiting diseaseassociated variants xx • Disease-associated SNPs enriched for enhancers in relevant cell types • E. g. lupus SNP in GM enhancer disrupts Ets 1 predicted activator

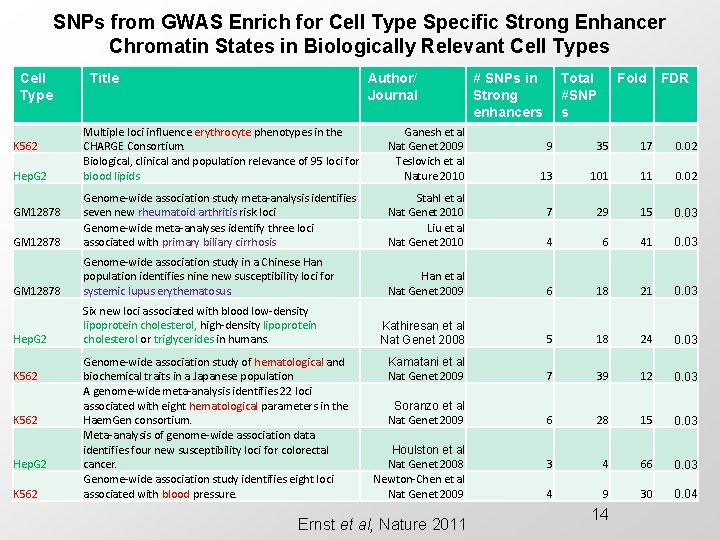
SNPs from GWAS Enrich for Cell Type Specific Strong Enhancer Chromatin States in Biologically Relevant Cell Types Cell Type Title Author/ Journal Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Biological, clinical and population relevance of 95 loci for blood lipids Ganesh et al Nat Genet 2009 Teslovich et al Nature 2010 GM 12878 Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis Stahl et al Nat Genet 2010 Liu et al Nat Genet 2010 GM 12878 Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Hep. G 2 Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. K 562 Hep. G 2 GM 12878 K 562 Hep. G 2 K 562 Genome-wide association study of hematological and biochemical traits in a Japanese population A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the Haem. Gen consortium. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Genome-wide association study identifies eight loci associated with blood pressure. # SNPs in Strong enhancers Total #SNP s Fold FDR 9 35 17 0. 02 13 101 11 0. 02 7 29 15 0. 03 4 6 41 0. 03 Han et al Nat Genet 2009 6 18 21 0. 03 Kathiresan et al Nat Genet 2008 5 18 24 0. 03 Kamatani et al Nat Genet 2009 7 39 12 0. 03 Soranzo et al Nat Genet 2009 6 28 15 0. 03 3 4 66 0. 03 4 9 30 0. 04 Houlston et al Nat Genet 2008 Newton-Chen et al Nat Genet 2009 Ernst et al, Nature 2011 14

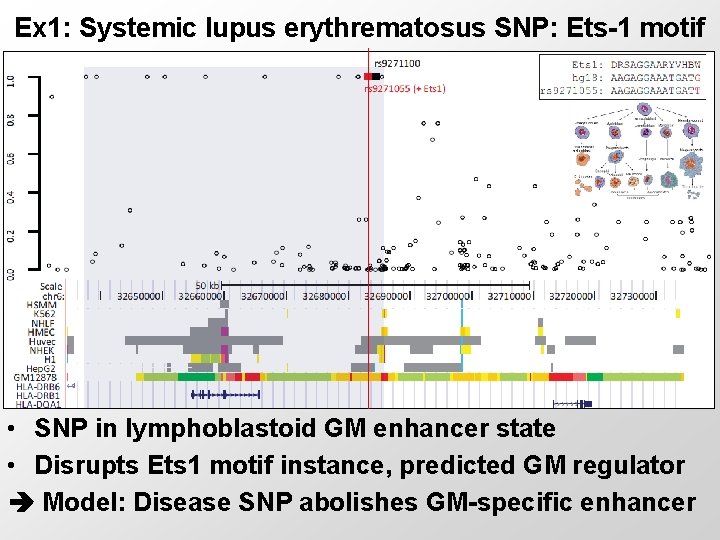
Ex 1: Systemic lupus erythrematosus SNP: Ets-1 motif • SNP in lymphoblastoid GM enhancer state • Disrupts Ets 1 motif instance, predicted GM regulator Model: Disease SNP abolishes GM-specific enhancer

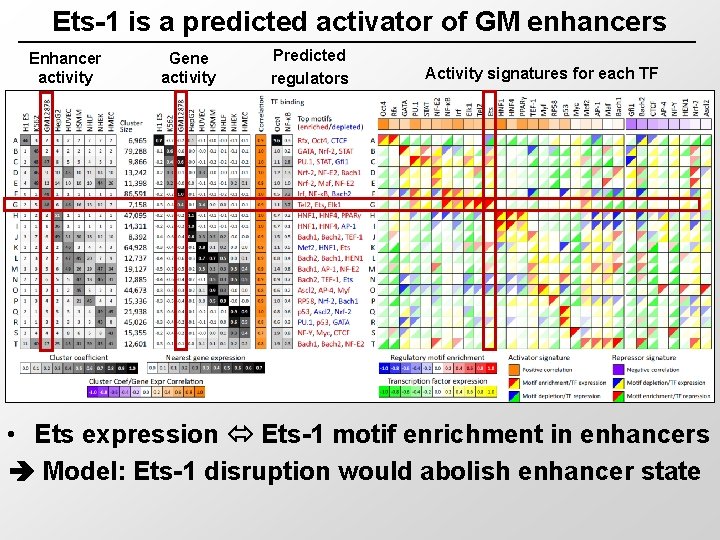
Ets-1 is a predicted activator of GM enhancers Enhancer activity Gene activity Predicted regulators Activity signatures for each TF • Ets expression Ets-1 motif enrichment in enhancers Model: Ets-1 disruption would abolish enhancer state

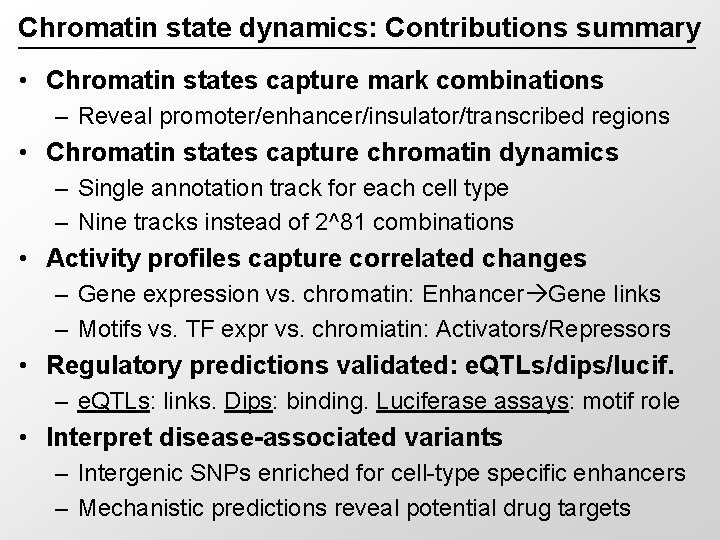
Chromatin state dynamics: Contributions summary • Chromatin states capture mark combinations – Reveal promoter/enhancer/insulator/transcribed regions • Chromatin states capture chromatin dynamics – Single annotation track for each cell type – Nine tracks instead of 2^81 combinations • Activity profiles capture correlated changes – Gene expression vs. chromatin: Enhancer Gene links – Motifs vs. TF expr vs. chromiatin: Activators/Repressors • Regulatory predictions validated: e. QTLs/dips/lucif. – e. QTLs: links. Dips: binding. Luciferase assays: motif role • Interpret disease-associated variants – Intergenic SNPs enriched for cell-type specific enhancers – Mechanistic predictions reveal potential drug targets

Collaborators and Acknowledgements MIT compbio group: • Pouya Kheradpour • Lucas Ward • Manolis Kellis ENCODE consortium Funding • NHGRI, NIH, NSF, HHMI, Sloan Foundation MGH Pathology/HHMI: • Tarjei Mikkelsen • Noam Shoresh • Charles B. Epstein • Xiaolan Zhang • Li Wang • Robyn Issner • Michael Coyne • Manching Ku • Timothy Durham • Bradley E. Bernstein