Chemical biology is a scientific discipline spanning the

Chemical biology is a scientific discipline spanning the fields of chemistry and biology that involves the application of chemical techniques and tools, often compounds produced through synthetic chemistry, to the study and manipulation of biological systems. • Proteomics (study of proteins/enzymes) • Glycobiology • Molecular sensing

Enzymes (History) • First discovered by Eduard Buchner in 1897 who observed that yeast extracts can ferment sugar to alcohol • This proved that fermentation was promoted by molecules that continued to function when removed from cells • The first enzyme to be purified and crystallized was urease in 1926; these crystals consisted entirely of protein • Later, pepsin, trypsin and other digestive proteins were isolated and determined to be purely protein as well

Enzymes Ø Ø Enzymes are the catalysts of nature.
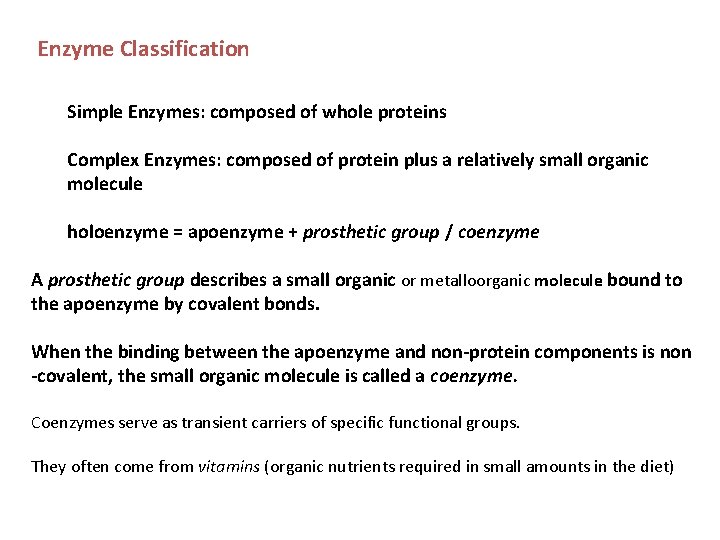
Enzyme Classification Simple Enzymes: composed of whole proteins Complex Enzymes: composed of protein plus a relatively small organic molecule holoenzyme = apoenzyme + prosthetic group / coenzyme A prosthetic group describes a small organic or metalloorganic molecule bound to the apoenzyme by covalent bonds. When the binding between the apoenzyme and non-protein components is non -covalent, the small organic molecule is called a coenzyme. Coenzymes serve as transient carriers of specific functional groups. They often come from vitamins (organic nutrients required in small amounts in the diet)
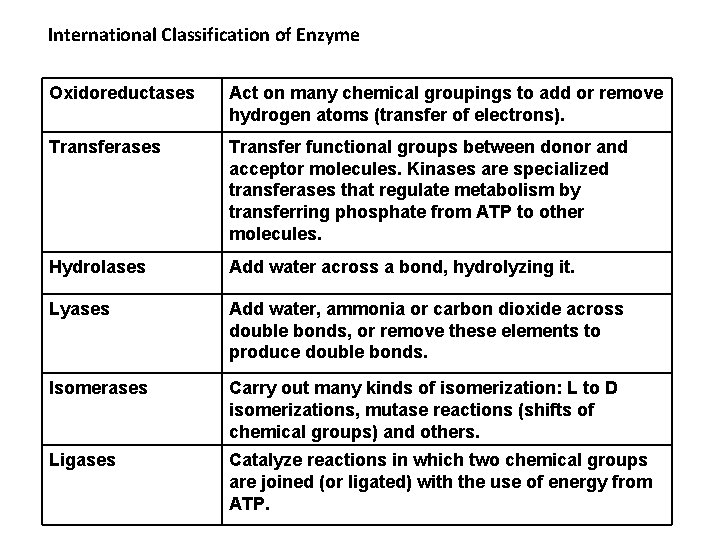
International Classification of Enzyme Oxidoreductases Act on many chemical groupings to add or remove hydrogen atoms (transfer of electrons). Transferases Transfer functional groups between donor and acceptor molecules. Kinases are specialized transferases that regulate metabolism by transferring phosphate from ATP to other molecules. Hydrolases Add water across a bond, hydrolyzing it. Lyases Add water, ammonia or carbon dioxide across double bonds, or remove these elements to produce double bonds. Isomerases Carry out many kinds of isomerization: L to D isomerizations, mutase reactions (shifts of chemical groups) and others. Ligases Catalyze reactions in which two chemical groups are joined (or ligated) with the use of energy from ATP.
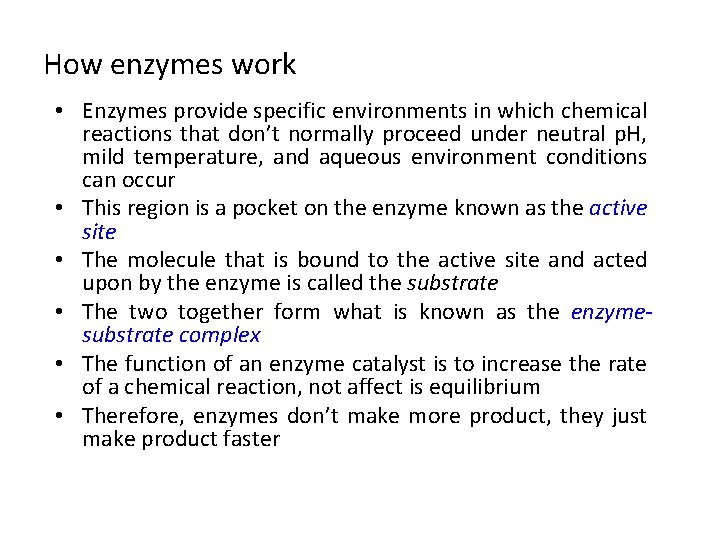
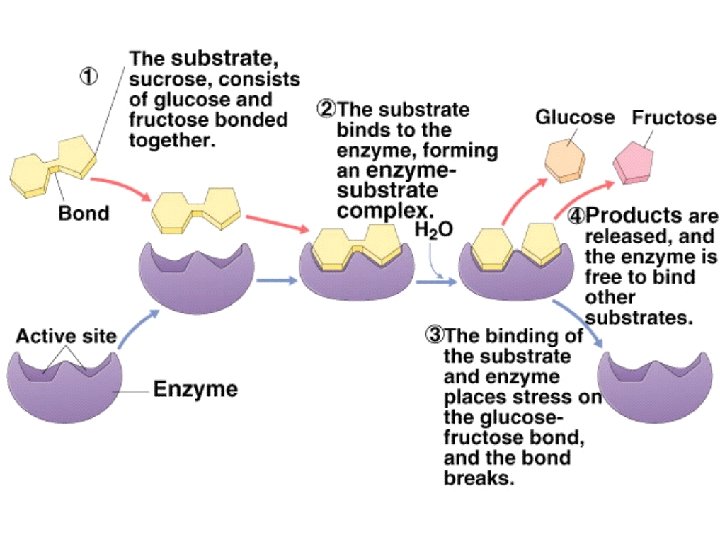
How enzymes work • Enzymes provide specific environments in which chemical reactions that don’t normally proceed under neutral p. H, mild temperature, and aqueous environment conditions can occur • This region is a pocket on the enzyme known as the active site • The molecule that is bound to the active site and acted upon by the enzyme is called the substrate • The two together form what is known as the enzymesubstrate complex • The function of an enzyme catalyst is to increase the rate of a chemical reaction, not affect is equilibrium • Therefore, enzymes don’t make more product, they just make product faster

Active Site • The area of an enzyme that binds to the substrate • Structure has a unique geometric shape that is designed to fit the molecular shape of the substrate • Each enzyme is substrate specific • Thus the active site that is complementary to the geometric shape of a substrate molecule • Active site is lined with residues and sometimes contains a co-factor • Active site residues have several important properties: – Charge [partial, dipoles, helix dipole] – p. Ka – Hydrophobicity – Flexibility – Reactivity
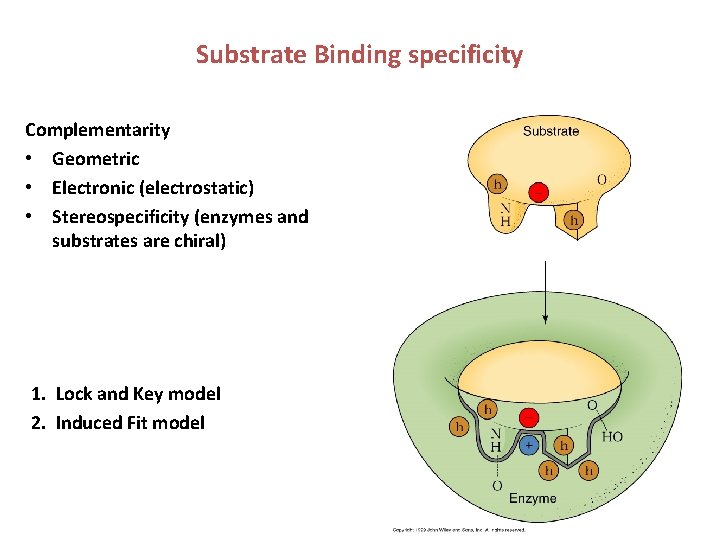
Substrate Binding specificity Complementarity • Geometric • Electronic (electrostatic) • Stereospecificity (enzymes and substrates are chiral) 1. Lock and Key model 2. Induced Fit model
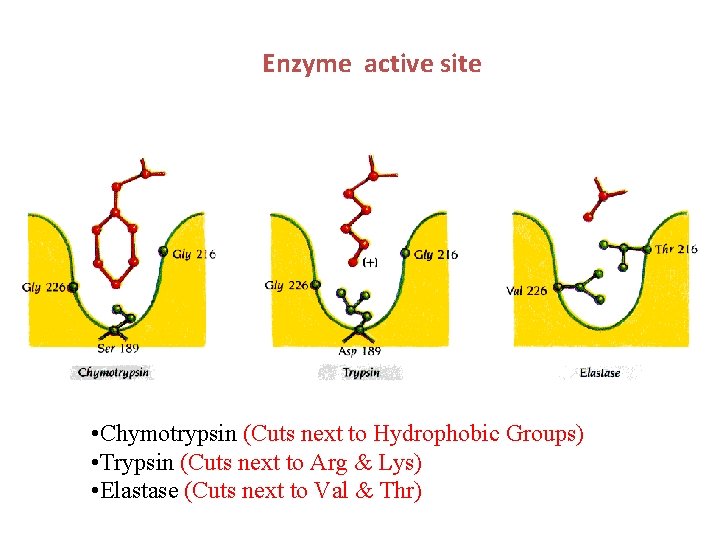
Enzyme active site • Chymotrypsin (Cuts next to Hydrophobic Groups) • Trypsin (Cuts next to Arg & Lys) • Elastase (Cuts next to Val & Thr)
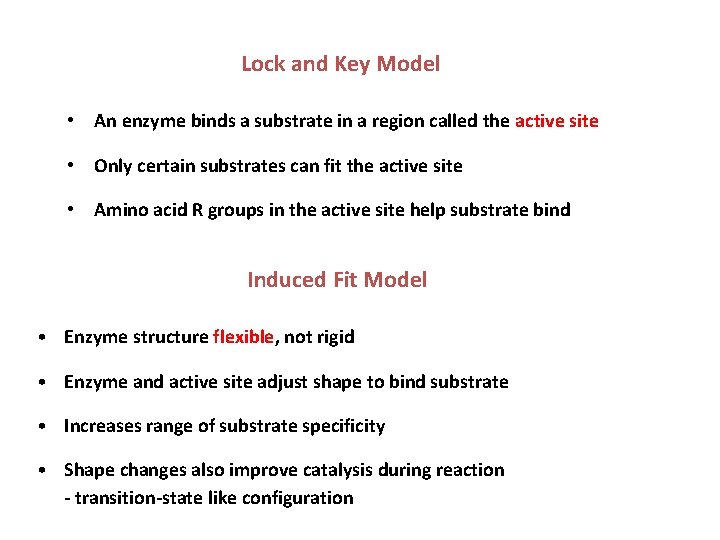
Lock and Key Model • An enzyme binds a substrate in a region called the active site • Only certain substrates can fit the active site • Amino acid R groups in the active site help substrate bind Induced Fit Model • Enzyme structure flexible, not rigid • Enzyme and active site adjust shape to bind substrate • Increases range of substrate specificity • Shape changes also improve catalysis during reaction - transition-state like configuration
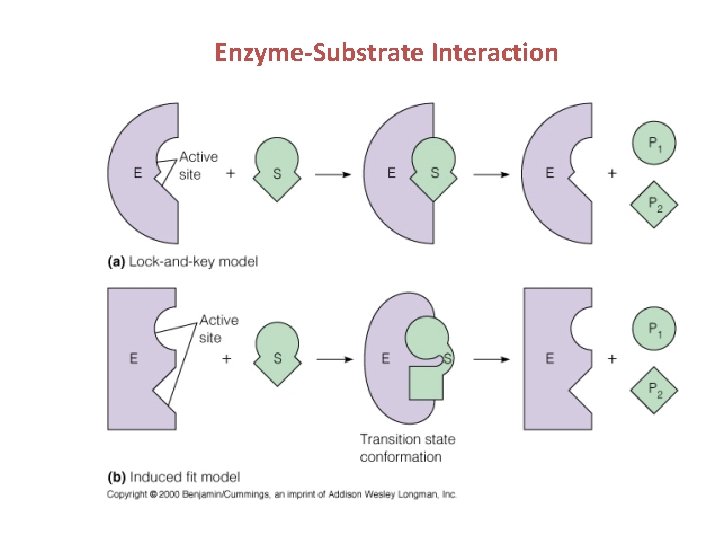
Enzyme-Substrate Interaction
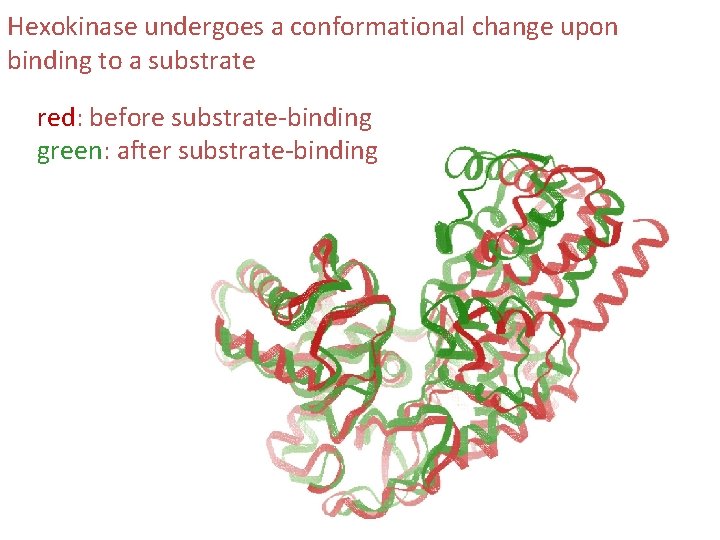
Hexokinase undergoes a conformational change upon binding to a substrate red: before substrate-binding green: after substrate-binding
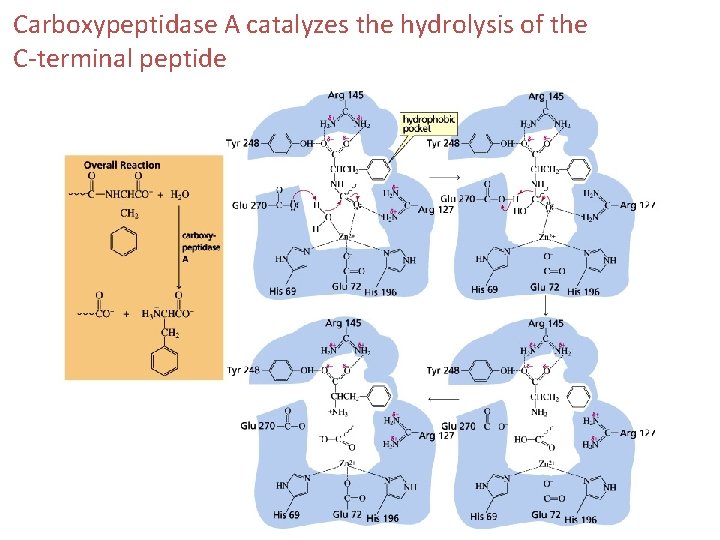
Carboxypeptidase A catalyzes the hydrolysis of the C-terminal peptide
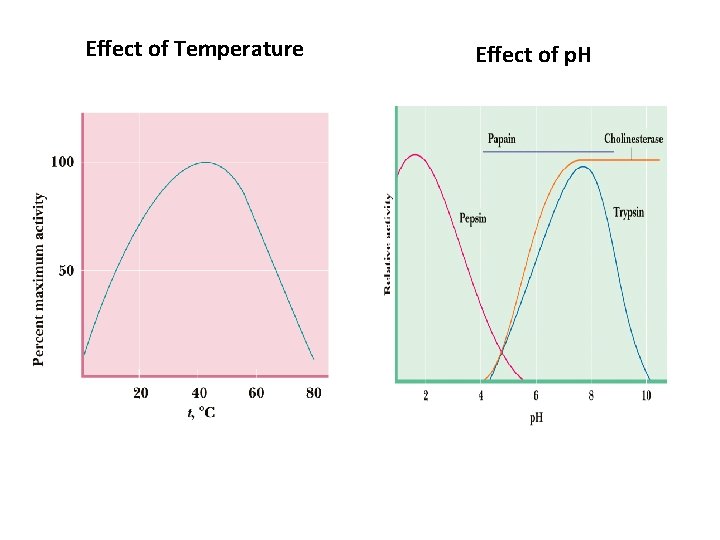
Effect of Temperature Effect of p. H
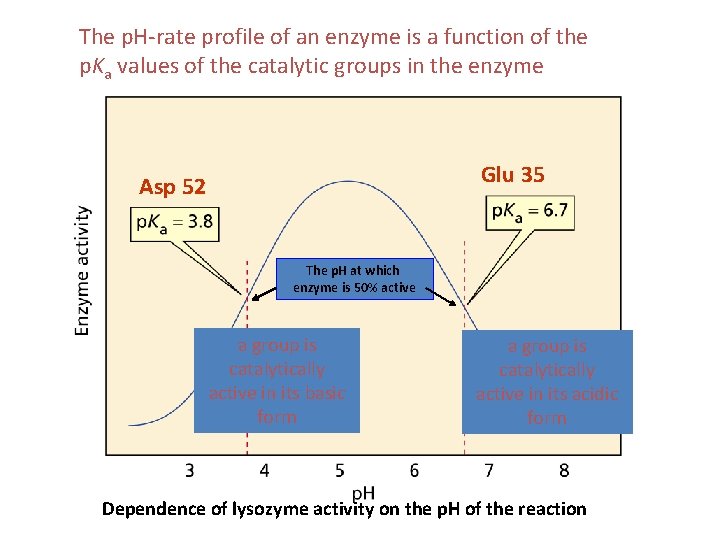
The p. H-rate profile of an enzyme is a function of the p. Ka values of the catalytic groups in the enzyme Glu 35 Asp 52 The p. H at which enzyme is 50% active a group is catalytically active in its basic form a group is catalytically active in its acidic form Dependence of lysozyme activity on the p. H of the reaction

The function of an enzyme catalyst is to increase the rate of a chemical reaction, not affect is equilibrium. Therefore, enzymes don’t make more product, they just make product faster.
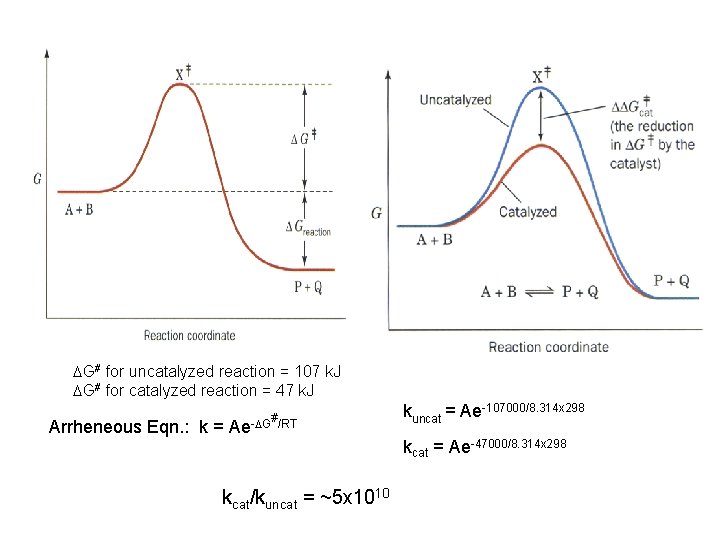
DG# for uncatalyzed reaction = 107 k. J DG# for catalyzed reaction = 47 k. J Arrheneous Eqn. : k = Ae -DG#/RT kcat/kuncat = ~5 x 1010 kuncat = Ae-107000/8. 314 x 298 kcat = Ae-47000/8. 314 x 298

How can an enzyme reduce the activation energy? (1) Binding to the substrate can be done such that the formation of the transition state is favored (2) Orientation and positioning of substrate(s) (3) Bonds in the substrate can be ‘activated’ by functional groups in the catalytic site
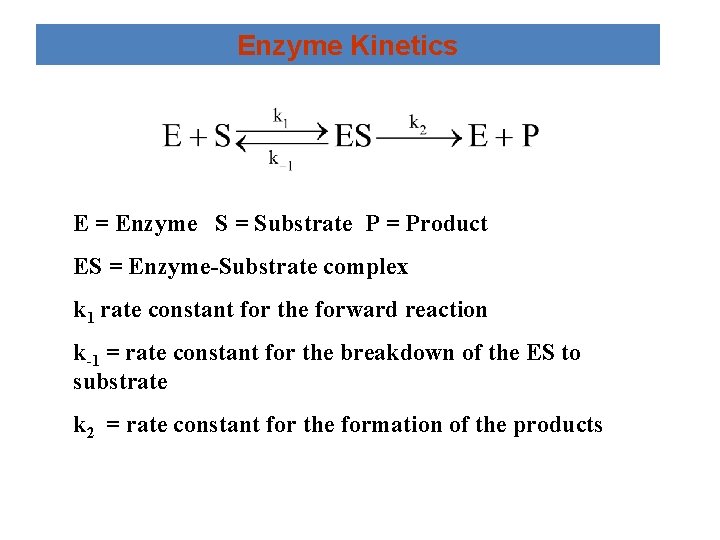
Enzyme Kinetics E = Enzyme S = Substrate P = Product ES = Enzyme-Substrate complex k 1 rate constant for the forward reaction k-1 = rate constant for the breakdown of the ES to substrate k 2 = rate constant for the formation of the products
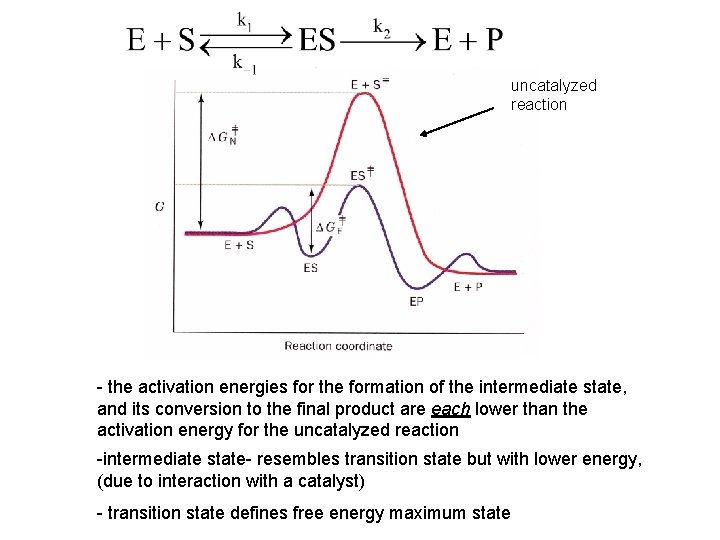
uncatalyzed reaction - the activation energies for the formation of the intermediate state, and its conversion to the final product are each lower than the activation energy for the uncatalyzed reaction -intermediate state- resembles transition state but with lower energy, (due to interaction with a catalyst) - transition state defines free energy maximum state
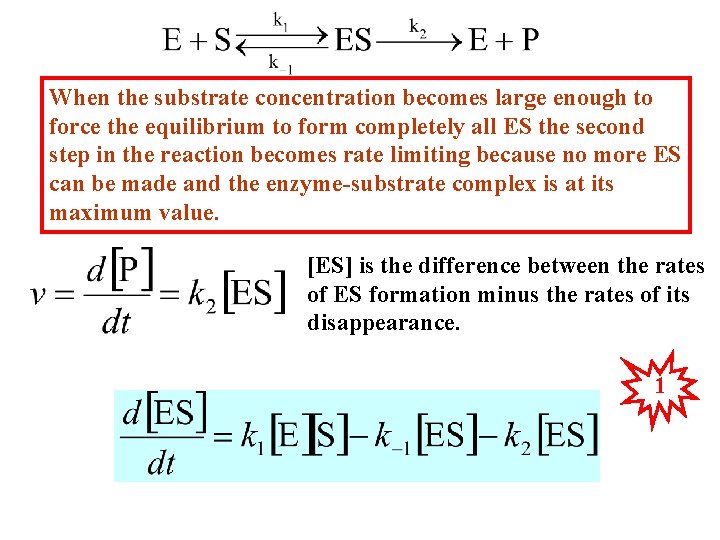
When the substrate concentration becomes large enough to force the equilibrium to form completely all ES the second step in the reaction becomes rate limiting because no more ES can be made and the enzyme-substrate complex is at its maximum value. [ES] is the difference between the rates of ES formation minus the rates of its disappearance. 1
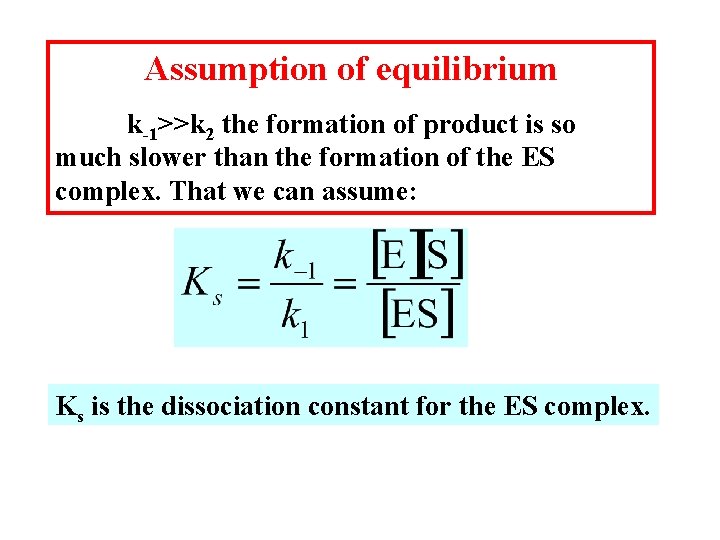
Assumption of equilibrium k-1>>k 2 the formation of product is so much slower than the formation of the ES complex. That we can assume: Ks is the dissociation constant for the ES complex.
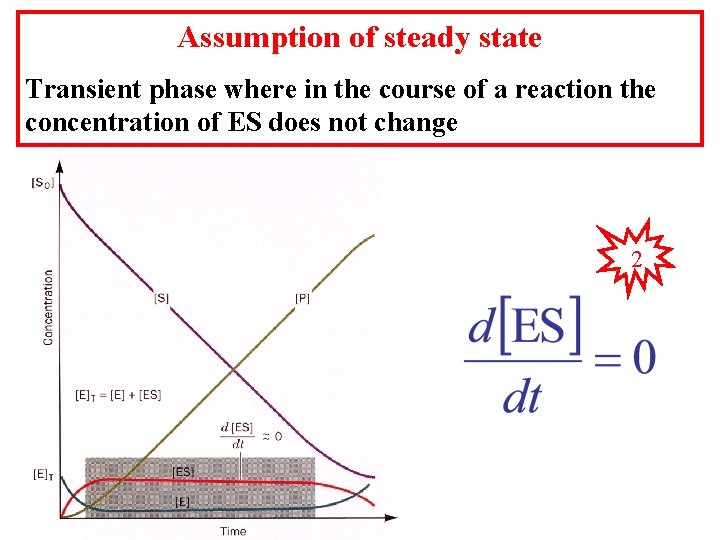
Assumption of steady state Transient phase where in the course of a reaction the concentration of ES does not change 2
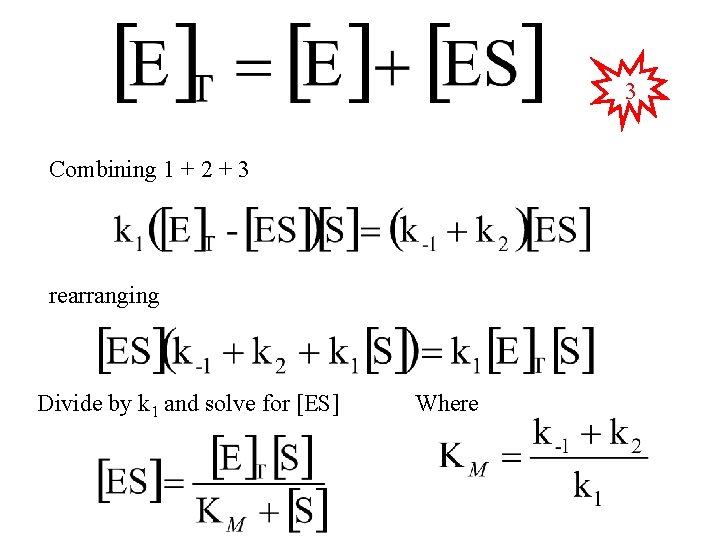
3 Combining 1 + 2 + 3 rearranging Divide by k 1 and solve for [ES] Where
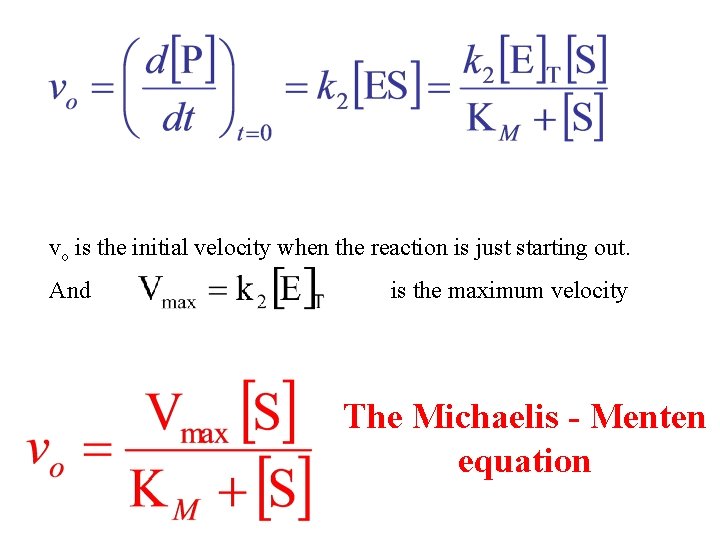
vo is the initial velocity when the reaction is just starting out. And is the maximum velocity The Michaelis - Menten equation
![Michaelis – Menten Kinetics low [S], v is proportional to [S] - first order Michaelis – Menten Kinetics low [S], v is proportional to [S] - first order](http://slidetodoc.com/presentation_image_h2/27d9f7ff187440b8ae2707245b3b0807/image-27.jpg)
Michaelis – Menten Kinetics low [S], v is proportional to [S] - first order high [S], v is independent of [S] - zero order The Km is the substrate concentration where vo equals one-half Vmax
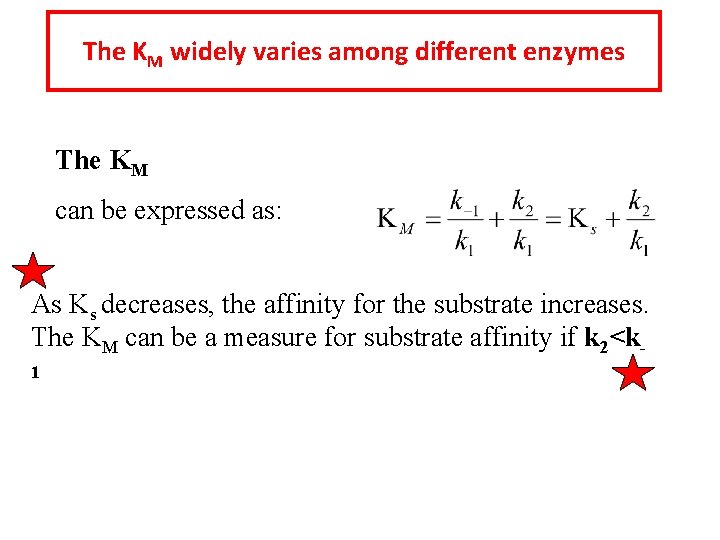
The KM widely varies among different enzymes The KM can be expressed as: As Ks decreases, the affinity for the substrate increases. The KM can be a measure for substrate affinity if k 2<k 1
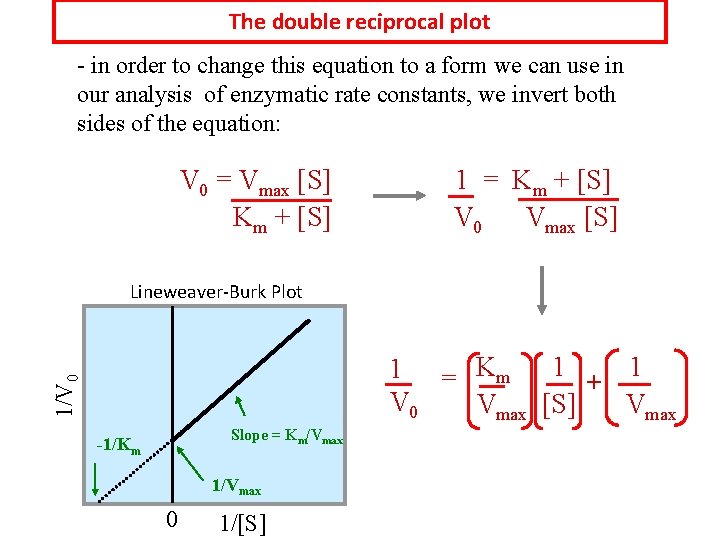
The double reciprocal plot - in order to change this equation to a form we can use in our analysis of enzymatic rate constants, we invert both sides of the equation: V 0 = Vmax [S] Km + [S] 1 = Km + [S] V 0 Vmax [S] Lineweaver-Burk Plot 1/V 0 1 1 1 = Km + V 0 Vmax [S] Vmax Slope = Km/Vmax -1/Km 1/Vmax 0 1/[S]
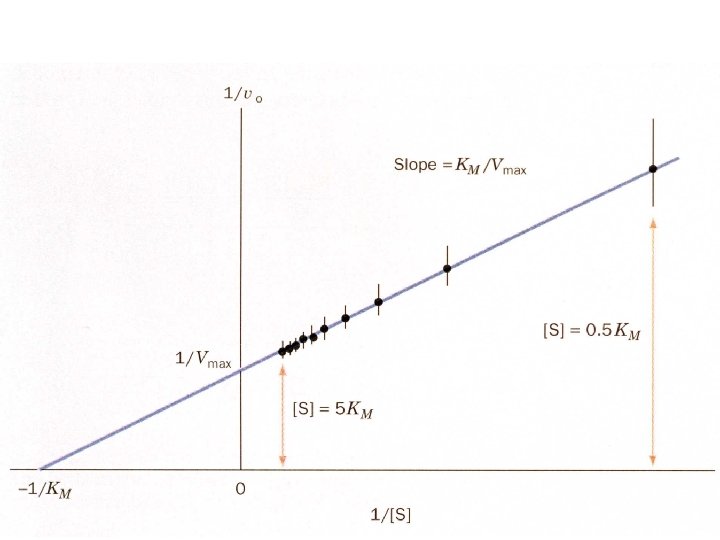
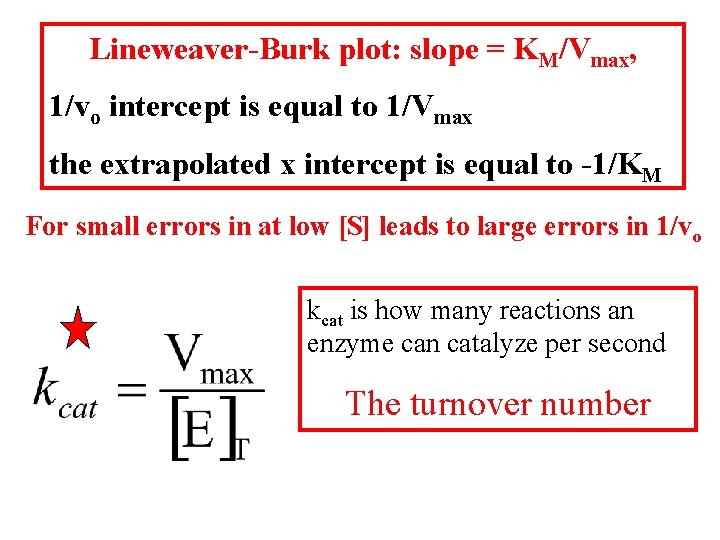
Lineweaver-Burk plot: slope = KM/Vmax, 1/vo intercept is equal to 1/Vmax the extrapolated x intercept is equal to -1/KM For small errors in at low [S] leads to large errors in 1/vo kcat is how many reactions an enzyme can catalyze per second The turnover number
![V 0 = Vmax [S] KM + [S] For Michaelis -Menton kinetics k 2= V 0 = Vmax [S] KM + [S] For Michaelis -Menton kinetics k 2=](http://slidetodoc.com/presentation_image_h2/27d9f7ff187440b8ae2707245b3b0807/image-32.jpg)
V 0 = Vmax [S] KM + [S] For Michaelis -Menton kinetics k 2= kcat When [S] << KM very little ES is formed and [E] = [E]T and kcat/KM is a measure of catalytic efficiency
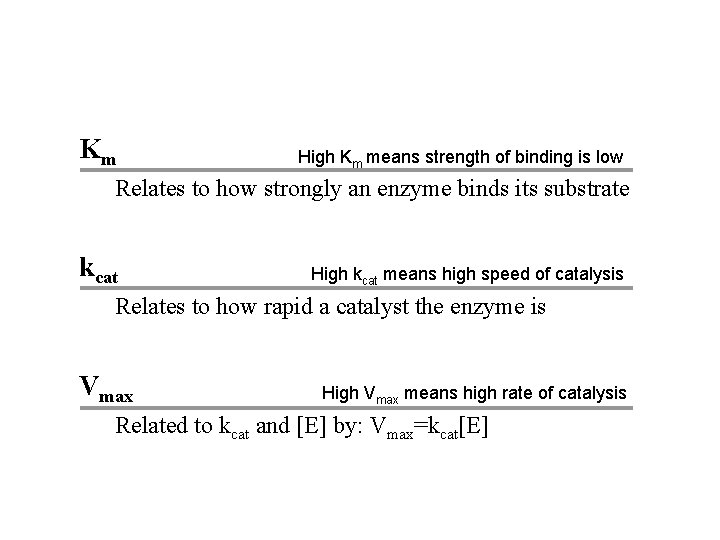
Km High Km means strength of binding is low Relates to how strongly an enzyme binds its substrate kcat High kcat means high speed of catalysis Relates to how rapid a catalyst the enzyme is Vmax High Vmax means high rate of catalysis Related to kcat and [E] by: Vmax=kcat[E]
![• kcat = turnover number; kcat = Vmax/[E]T • kcat/Km is a measure • kcat = turnover number; kcat = Vmax/[E]T • kcat/Km is a measure](http://slidetodoc.com/presentation_image_h2/27d9f7ff187440b8ae2707245b3b0807/image-34.jpg)
• kcat = turnover number; kcat = Vmax/[E]T • kcat/Km is a measure of activity, catalytic efficiency KM is a useful indicator of the affinity of an enzyme for the substrate A low KM indicates a high affinity for the substrate A high kcat/KM ratio implies an efficient enzyme This could result from: Large kcat Small KM
Enzyme Inhibition • Inhibitors: compounds that decrease activity of the enzyme • Can decrease binding of substrate (affect KM), or turnover # (affect kcat) or both • Most drugs are enzyme inhibitors • Inhibitors are also important for determining enzyme mechanisms and the nature of the active site. • Important to know how inhibitors work – facilitates drug design, inhibitor design. • Antibiotics inhibit enzymes by affecting bacterial metabolism • Nerve Gases cause irreversible enzyme inhibition • Insecticides – choline esterase inhibitors • Many heavy metal poisons work by irreversibly inhibiting enzymes, especially cysteine residues

Types of Enzyme Inhibition • Reversible inhibition reversibly bind and dissociate from enzyme, activity of enzyme recovered on removal of inhibitor - usually non-covalent in nature – Competitive – Uncompetitive – Noncompetitive (Mixed) • Irreversible inhibition inactivators that irreversibly associate with enzyme activity of enzyme not recovered on removal - usually covalent in nature
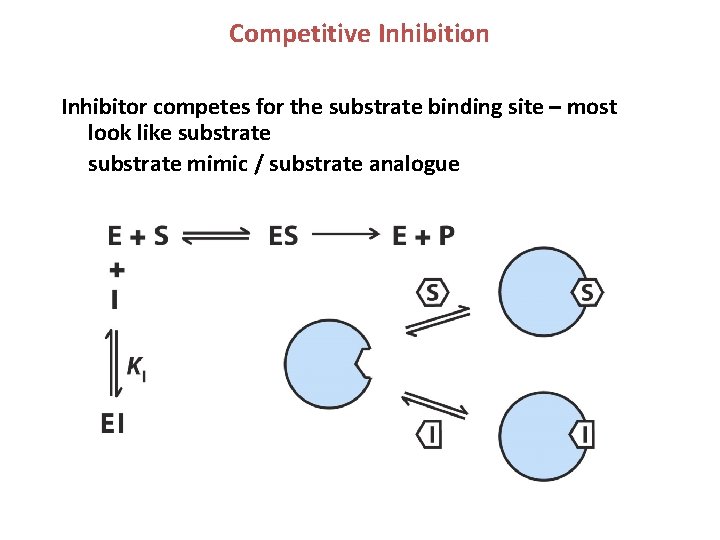
Competitive Inhibition Inhibitor competes for the substrate binding site – most look like substrate mimic / substrate analogue
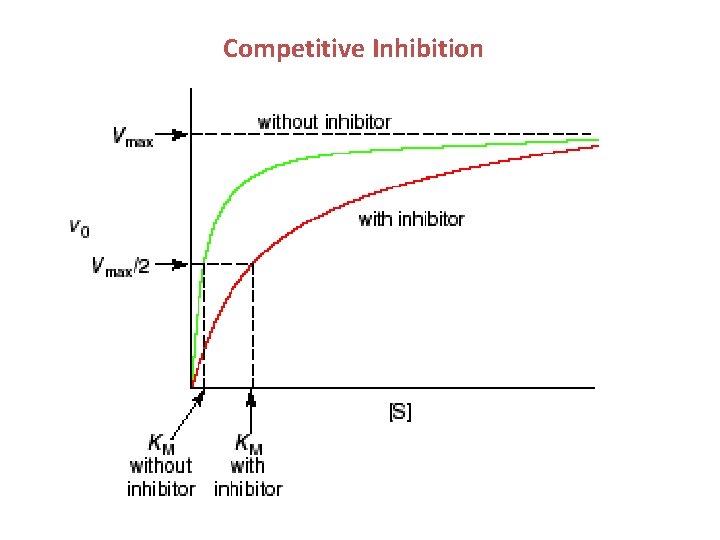
Competitive Inhibition
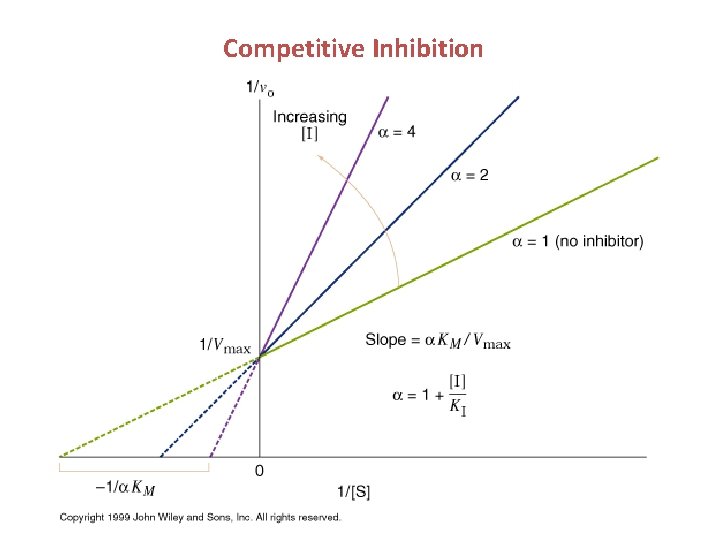
Competitive Inhibition
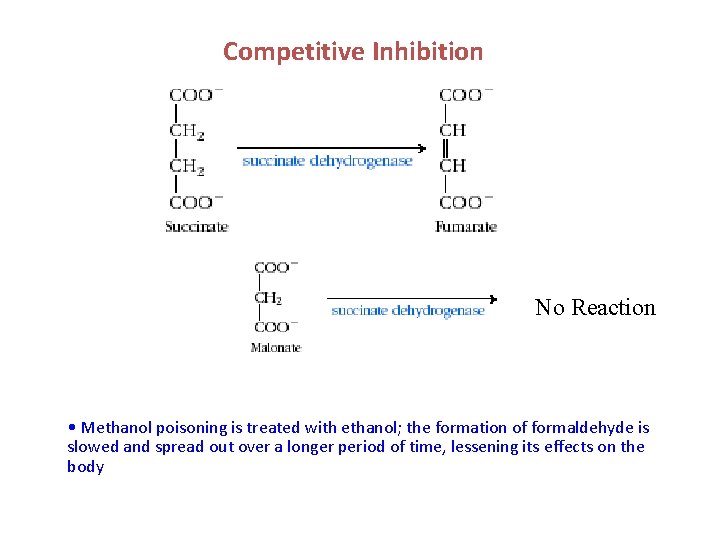
Competitive Inhibition No Reaction • Methanol poisoning is treated with ethanol; the formation of formaldehyde is slowed and spread out over a longer period of time, lessening its effects on the body
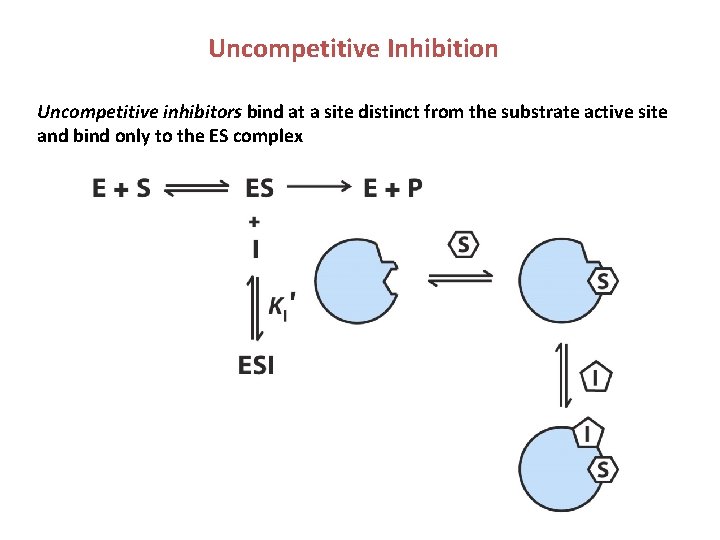
Uncompetitive Inhibition Uncompetitive inhibitors bind at a site distinct from the substrate active site and bind only to the ES complex
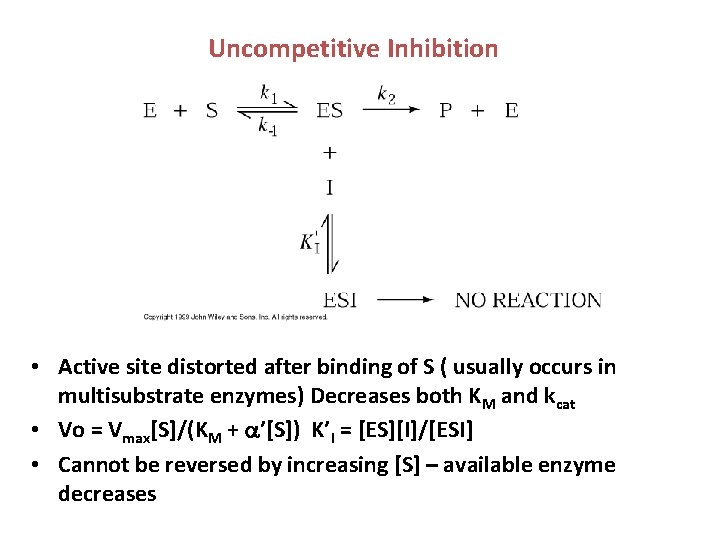
Uncompetitive Inhibition • Active site distorted after binding of S ( usually occurs in multisubstrate enzymes) Decreases both KM and kcat • Vo = Vmax[S]/(KM + ’[S]) K’I = [ES][I]/[ESI] • Cannot be reversed by increasing [S] – available enzyme decreases
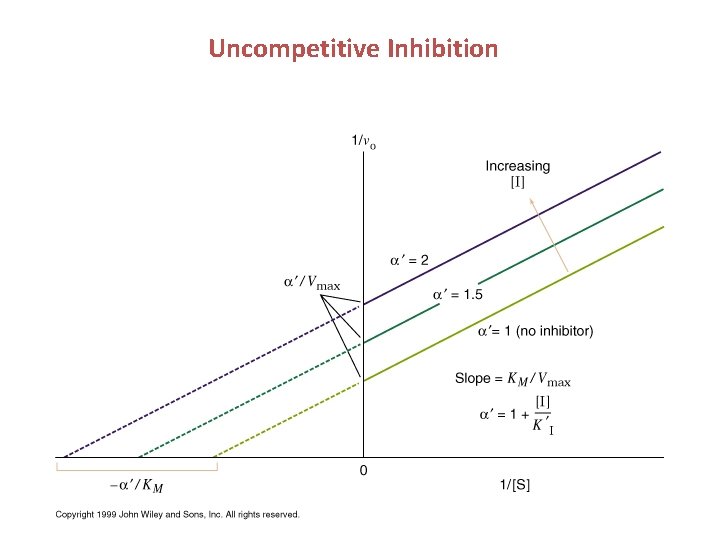
Uncompetitive Inhibition
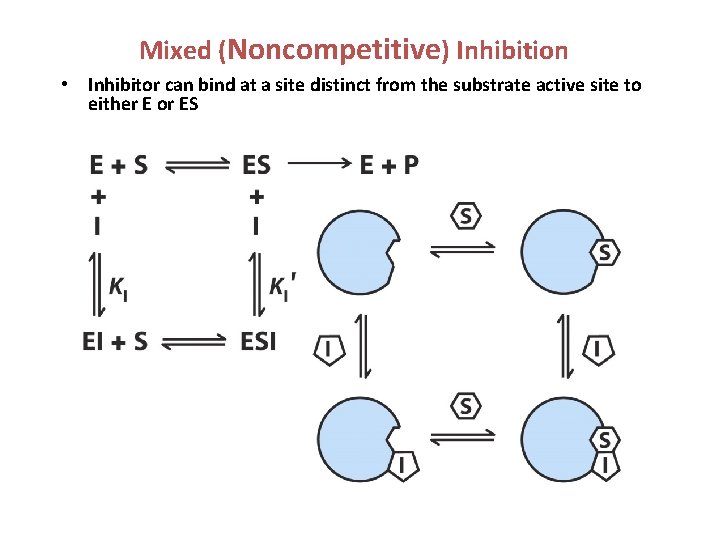
Mixed (Noncompetitive) Inhibition • Inhibitor can bind at a site distinct from the substrate active site to either E or ES
![Mixed Inhibition • Vo = Vmax[S]/( KM + ’[S]) • Vmax decreases; KM can Mixed Inhibition • Vo = Vmax[S]/( KM + ’[S]) • Vmax decreases; KM can](http://slidetodoc.com/presentation_image_h2/27d9f7ff187440b8ae2707245b3b0807/image-45.jpg)
Mixed Inhibition • Vo = Vmax[S]/( KM + ’[S]) • Vmax decreases; KM can go up or down.

Non-competitive inhibition models a system where the inhibitor and the substrate may both be bound to the enzyme at any given time. When both the substrate and the inhibitor are bound, the enzyme-substrate-inhibitor complex cannot form product and can only be converted back to the enzyme-substrate complex or the enzyme-inhibitor complex. Non-competitive inhibition is distinguished from general mixed inhibition in that the inhibitor has an equal affinity for the enzyme and the enzyme-substrate complex. Mixed inhibition refers to a combination of two different types of reversible enzyme inhibition – competitive inhibition and uncompetitive inhibition. The term 'mixed' is used when the inhibitor can bind to either the free enzyme or the enzyme-substrate complex. In mixed inhibition, the inhibitor binds to a site different from the active site where the substrate binds. Mixed inhibition results in a decrease in the apparent affinity of the enzyme for the substrate ( Kmapp > Km, a decrease in apparent affinity means the Km value appears to increase) and a decrease in the apparent maximum enzyme reaction rate (Vmaxapp < Vmax). Mathematically, mixed inhibition occurs when the factors α and α’ (introduced into the Michaelis. Menten equation to account for competitive and uncompetitive inhibition, respectively) are both greater than 1. In the special case where α = α’, noncompetitive inhibition occurs, in which case Vmaxapp is reduced but Km is unaffected. This is very unusual in practice
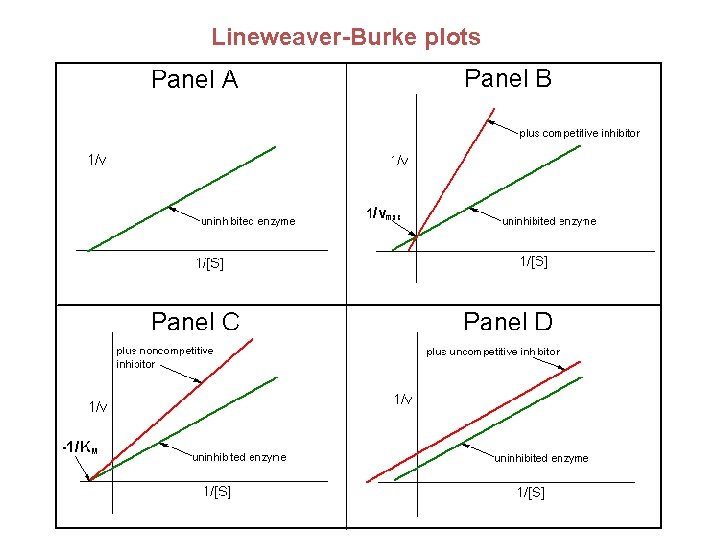
Lineweaver-Burke plots
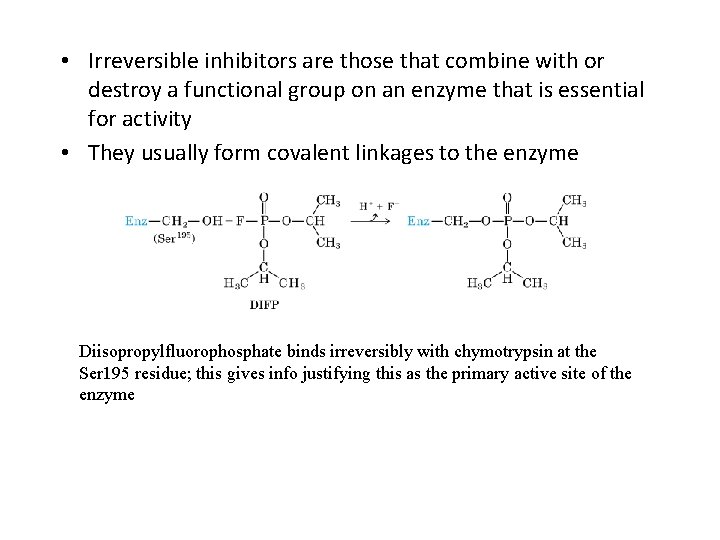
• Irreversible inhibitors are those that combine with or destroy a functional group on an enzyme that is essential for activity • They usually form covalent linkages to the enzyme Diisopropylfluorophosphate binds irreversibly with chymotrypsin at the Ser 195 residue; this gives info justifying this as the primary active site of the enzyme

• A special class of irreversible inhibitors is the suicide inactivators • These are unreactive until bound to the active site • They are designed to carry out the first few steps of a normal enzyme reaction, but instead of forming a product, they form a highly reactive compound that binds irreversibly to the enzyme • They are sometimes called mechanism-based inactivators, because they use the normal enzyme mechanism to lead to the inactivation • These are often used in drug design
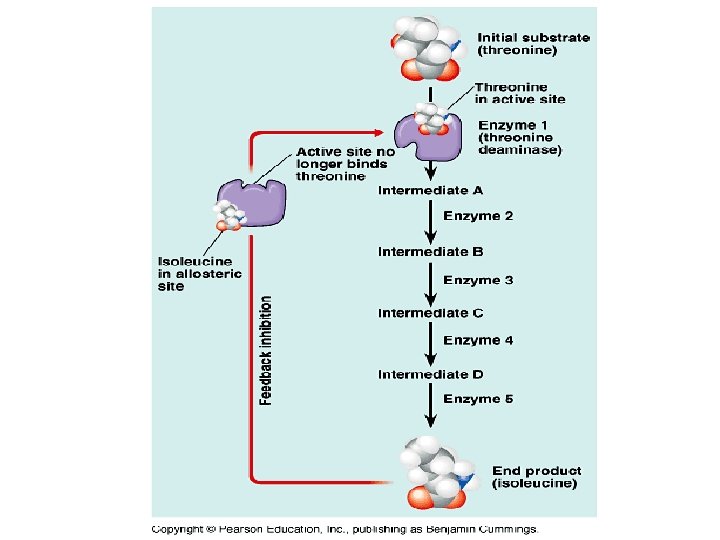
Regulatory Enzymes • These are enzymes that set the rate of a metabolic pathway by catalyzing the slowest or rate-limiting reaction • They experience increased or decreased catalytic activity in response to certain external signals • There are two major classes of regulatory enzymes in metabolic pathways • Allosteric enzymes bind regulatory compounds called allosteric modulators reversible, noncovalent interactions • Others regulate by reversible covalent modification • In some pathways, the regulatory enzyme is inhibited by the end product of the pathway whenever the end product concentration exceeds the cell’s requirement • When the regulatory enzyme is slowed, all subsequent enzymes operate at reduced rates • This is known as feedback inhibition
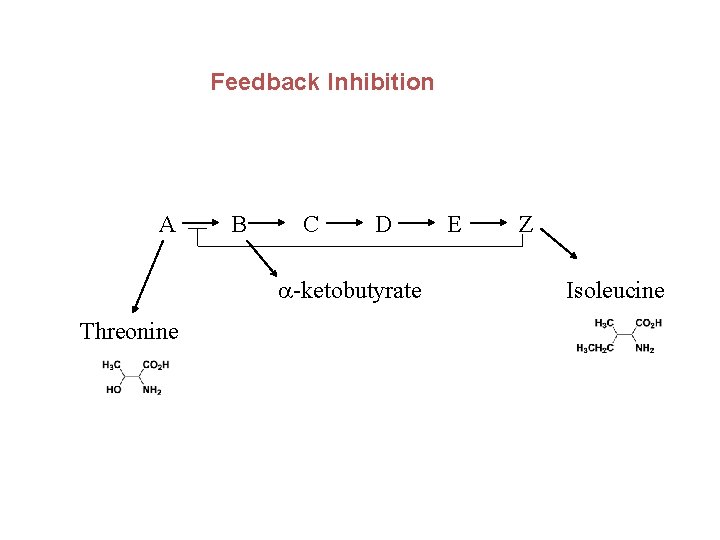
Feedback Inhibition A B C D a-ketobutyrate Threonine E Z Isoleucine
Allosteric regulation When a small molecule can act as an effector or regulator to activate or inactivate an action of a protein - the protein is said to be under allosteric control. The binding of the small ligand is distant from the protein’s active site and regulation is a result of a conformational change in the protein when the ligand is bound Many types of proteins show allosteric control: - haemoglobin (NOT myoglobin) - various enzymes - various gene-regulating proteins
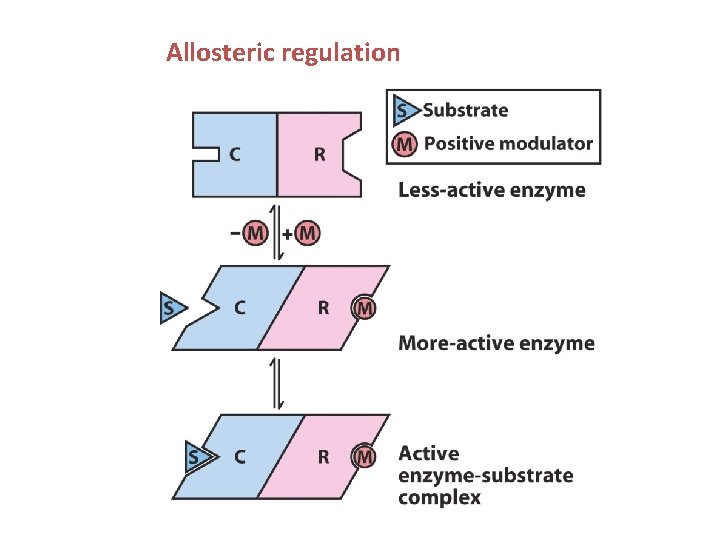
Allosteric regulation

Example: Phosphofructokinase and ATP Substrate: Fructose-6 -phosphate Reaction phosphofructokinase fructose-6 -phosphate + ATP fructose-1, 6 -bisphosphate + ADP © 2008 Paul Billiet ODWS
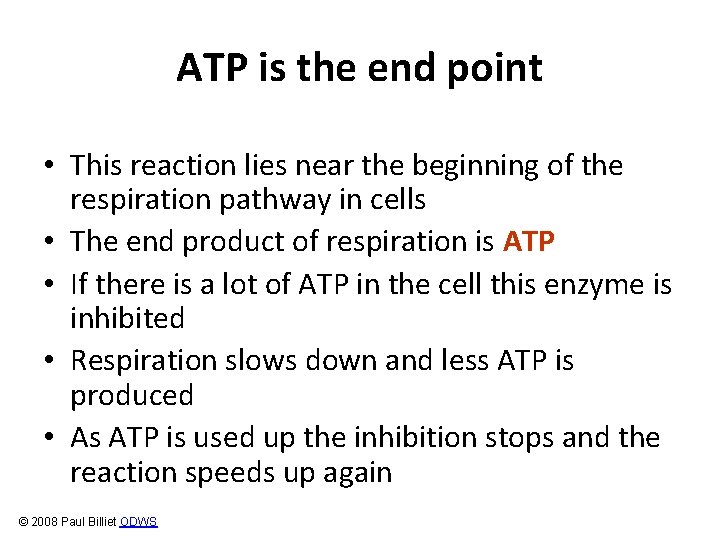
ATP is the end point • This reaction lies near the beginning of the respiration pathway in cells • The end product of respiration is ATP • If there is a lot of ATP in the cell this enzyme is inhibited • Respiration slows down and less ATP is produced • As ATP is used up the inhibition stops and the reaction speeds up again © 2008 Paul Billiet ODWS
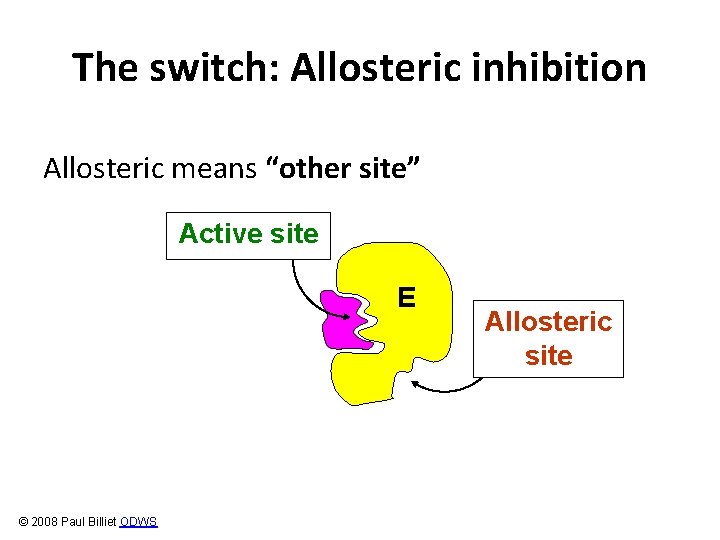
The switch: Allosteric inhibition Allosteric means “other site” Active site E © 2008 Paul Billiet ODWS Allosteric site
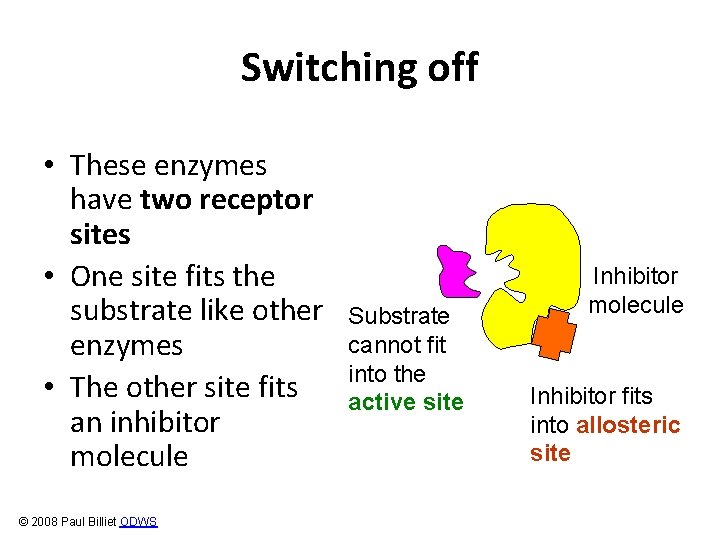
Switching off • These enzymes have two receptor sites • One site fits the substrate like other enzymes • The other site fits an inhibitor molecule © 2008 Paul Billiet ODWS Substrate cannot fit into the active site Inhibitor molecule Inhibitor fits into allosteric site
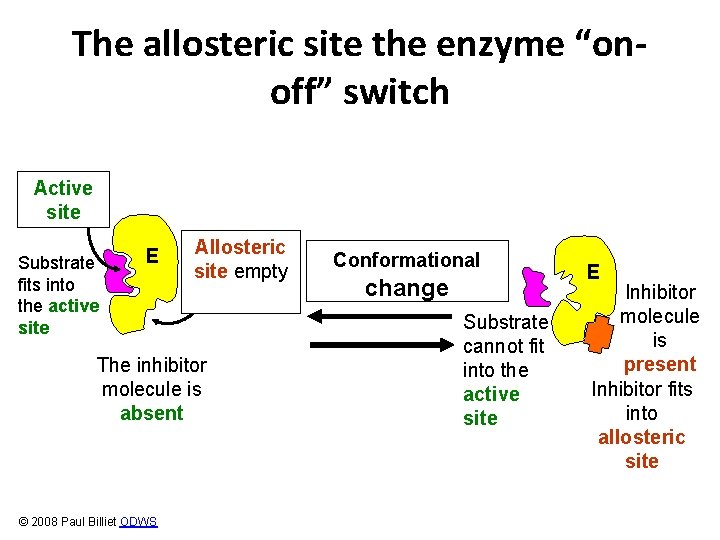
The allosteric site the enzyme “onoff” switch Active site Substrate fits into the active site E Allosteric site empty The inhibitor molecule is absent © 2008 Paul Billiet ODWS Conformational change Substrate cannot fit into the active site E Inhibitor molecule is present Inhibitor fits into allosteric site
A change in shape • When the inhibitor is present it fits into its site and there is a conformational change in the enzyme molecule • The enzyme’s molecular shape changes • The active site of the substrate changes • The substrate cannot bind with the substrate © 2008 Paul Billiet ODWS
- Slides: 60