Characterization of RDR Gene Expression Johnny R Nunez
- Slides: 1

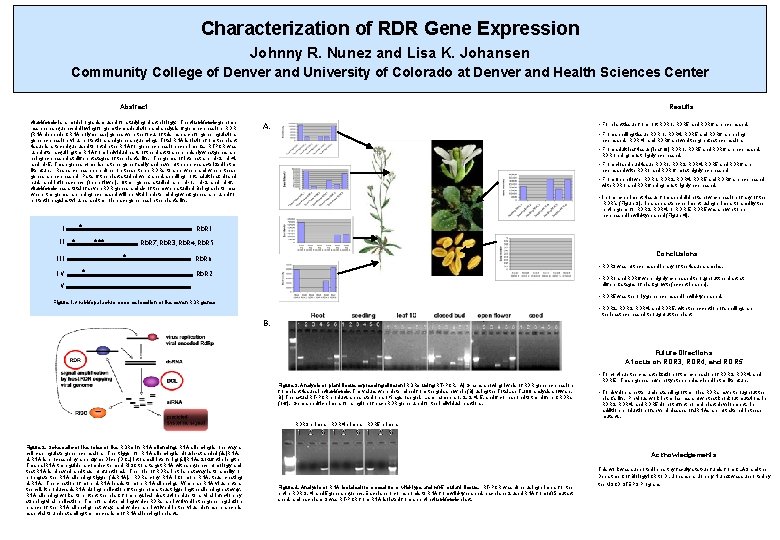
Characterization of RDR Gene Expression Johnny R. Nunez and Lisa K. Johansen Community College of Denver and University of Colorado at Denver and Health Sciences Center Abstract Results Arabidopsis is a model organism used for studying plant biology. The Arabidopsis genome has been sequenced allowing for genetic manipulation and analysis of gene expression. RDR (RNA dependent RNA polymerase) genes were the focus of this research for gene regulation, gene expression, virus protection, and gene sequencing. Total RNA isolation from the plant tissue is a technique used to obtain the RNA for gene expression experiments. RTPCR was used after acquiring the RNA from individual parts of the plant to see precisely what genes are being expressed at different stages of the plant’s life. The genes of interest are rdr-3, rdr-4, and rdr-5. These genes comprise a three gene family and have not been characterized in the literature. Research has been done on these three RDRs to see when and where these genes are expressed. Parts of the plant studied were: seed, seedling, root, adult leaf, closed bud, and inflorescence (open flower). Other genes studied are rdr-1, rdr-2 and rdr-6. Arabidopsis has a total of seven RDR genes and six of them were studied. Being able to map where the genes are being expressed will be vital in determining what genes are used for protection against viruses and the role each gene has in the plant’s life. * I II * IV expressed. RDR 4 and RDR 6 showed the greatest expression. • From adult leaf tissue (leaf 10) RDR 1, RDR 5 and RDR 6 are expressed. RDR 1 being most highly expressed. • From closed bud tissue RDR 1, RDR 2, RDR 4, RDR 5 and RDR 6 are expressed with RDR 1 and RDR 6 most highly expressed. • From Open flower RDR 1, RDR 2, RDR 4, RDR 5 and RDR 6 are expressed with RDR 1 and RDR 6 being most highly expressed. • In one experiment, tissue from seed did not show expression of any of the RDR’s (Figure 3). In a separate experiment, using primers to amplify the entire gene for RDR 3, RDR 4, or RDR 5, RDR 5 was shown to be expressed in wild-type seed (Figure 4). RDR 7, RDR 3, RDR 4, RDR 5 * * • From seedling tissue RDR 1, RDR 4, RDR 5 and RDR 6 are being RDR 1 *** III • For plant tissue from root RDR 1, RDR 5 and RDR 6 are expressed. A. Conclusions RDR 6 • RDR 3 was not expressed in any of the tissue samples. RDR 2 • RDR 1 and RDR 6 were highly expressed throughout the plant at V different stages of plant growth (except in seed). • RDR 5 was the only gene expressed in wild-type seed. Figure 1. Arabidopsis chromosome location of the seven RDR genes • RDR 2, RDR 3, RDR 4, and RDR 5, with the exception of seedling, are the least expressed throughout the plant. B. Future Directions A focus on RDR 3, RDR 4, and RDR 5 • To continue the characterization of the expression of RDR 3, RDR 4, and Figure 3. Analysis of plant tissue expressing different RDRs using RT-PCR. A) Graphs showing levels of RDR gene expression from plant tissues in Arabidopsis. The values were determined from the gels shown in (B) using the Total. Lab TL 100 analysis software. B) The actual RT-PCR products separated on a 1% agarose gel. Lane numbers 1, 2, 3, 4, 5, and 6 correspond to the different RDRs (1 -6). Gene specific primers for a region of each RDR gene used for the individual reactions. RDR 5. These genes have not yet been described in the literature. • To develop a better understanding of the roles RDRs have throughout the plant’s life. Previous work in the lab has shown that knockout mutations in RDR 3, RDR 4, and RDR 5 do not affect normal plant development. In addition, production of several classes of si. RNAs are not altered in these mutants. RDR 3 primers RDR 4 primers RDR 5 primers Figure 2. Schematic of the roles of the RDRs in RNA silencing is one way a cell can regulate gene expression. The trigger for RNA silencing is double-stranded (ds)RNA. ds. RNA is processed by an enzyme Dicer (DCL) into small interfering (si)RNAs 21 -26 nt in length. These si. RNA then guide a complex termed RISC to a target RNA with sequence homology and that RNA is cleaved and thus nonfunctional. The role of RDRs in this pathway is to amplify or propagate the RNA silencing trigger (ds. RNA). RDRs copy RNA into more RNA, thus creating ds. RNA. The creation of more ds. RNA leads to more RNA silencing. When an RNA virus enters the cell, it produces ds. RNA during replication of the genome, thus triggering the silencing pathway. RNA silencing works to protect the plant from physical destruction due to a viral infection by stopping viral replication. Therefore, determining which RDRs are involved in the gene regulation branch of the RNA silencing pathway, and which are involved in the virus defense branch is essential to understanding the mechanism of RNA silencing in plants. Acknowledgements Figure 4. Analysis of RNA isolated from seed from wild-type and rdr 5 mutant tissue. RT-PCR was done using primers for the entire RDR 3, 4, and 5 gene sequence. Each lane 1 corresponds to RNA from wild-type seed, each lane 2 used RNA from rdr 5 mutant seed, and each lane 3 was RT-PCR from RNA isolated from an entire Arabidopsis plant. This work was supported in part by faculty start-up funds from CLAS and the Department of Biology ICR to Dr. Johansen. Johnny Nunez was supported by the MSCD STEPS Program.

