CHAPTER DNA Replication Semiconservative DNA Replication 1 Watson




- Slides: 26

CHAPTER DNA Replication

Semiconservative DNA Replication • 1. Watson and Crick DNA model implies a mechanism for replication: • a. Unwind the DNA molecule. • b. Separate the two strands. • c. Make a complementary copy for each strand.

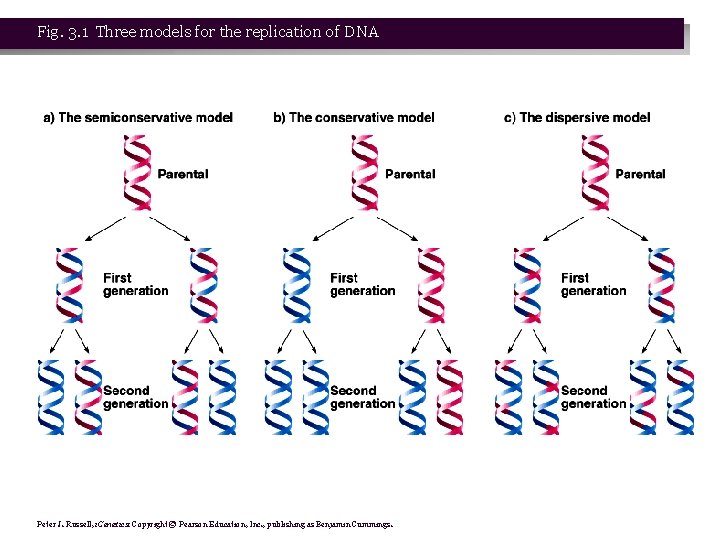
• 2. Three possible models were proposed for DNA replication: • a. Conservative model proposed both strands of one copy would be entirely old DNA, while the other copy would have both strands of new DNA. • b. Dispersive model was that ds. DNA might fragment, replicate ds. DNA, and then reassemble, creating a mosaic of old and new ds. DNA regions in each new chromosome. • c. Semiconservative model is that DNA strands separate, and a complementary strand is synthesized for each, so that sibling chromatids have one old and one new strand. This model was the winner in the Meselson and Stahl experiment.

Fig. 3. 1 Three models for the replication of DNA Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

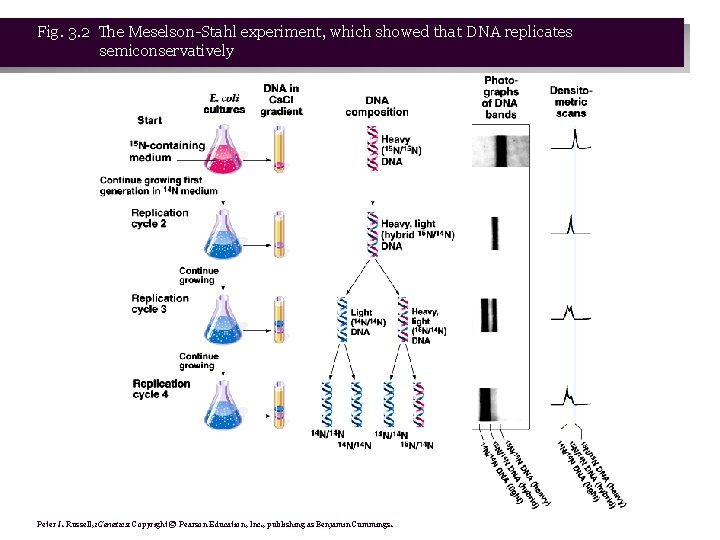
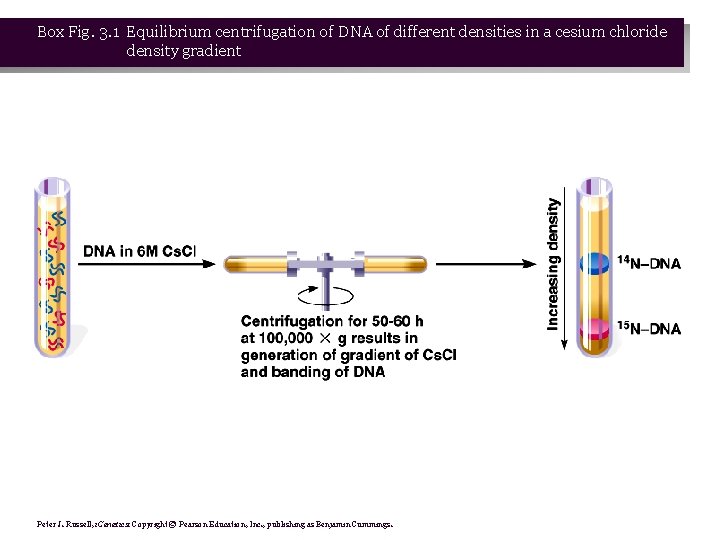
The Meselson-Stahl Experiment • 1. Meselson and Stahl (1958) grew E. coli in a heavy (not radioactive) isotope of nitrogen, 15 N in the form of 15 NH Cl. Because it is heavier, DNA containing 15 N is 4 more dense than DNA with normal 14 N, and so can be separated by Cs. Cl density gradient centrifugation (Box 3. 1). • 2. Once the E. coli were labeled with heavy 15 N, the researchers shifted the cells to medium containing normal 14 N, and took samples at time points. DNA was extracted from each sample and analyzed in Cs. Cl density gradients (Figure 3. 2).

Fig. 3. 2 The Meselson-Stahl experiment, which showed that DNA replicates semiconservatively Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

Box Fig. 3. 1 Equilibrium centrifugation of DNA of different densities in a cesium chloride density gradient Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

• 3. After one replication cycle in normal 14 N medium, all DNA had density intermediate between heavy and normal. After two replication cycles, there were two bands in the density gradient, one at the intermediate position, and one at the position for DNA containing entirely 14 N. • 4. Results compared with the three proposed models: • a. Does not fit conservative model, because after one generation there is a single intermediate band, rather than one with entirely 15 N DNA and another with entirely 14 N DNA. • b. The dispersive model predicted that a single band of DNA of intermediate density would be present in each generation, gradually becoming less dense as increasing amounts of 14 N were incorporated with each round of replication. Instead, Meselson and Stahl observed two bands of DNA, with the intermediate form decreasing over time. • c. The semiconservative model fits the data very well.

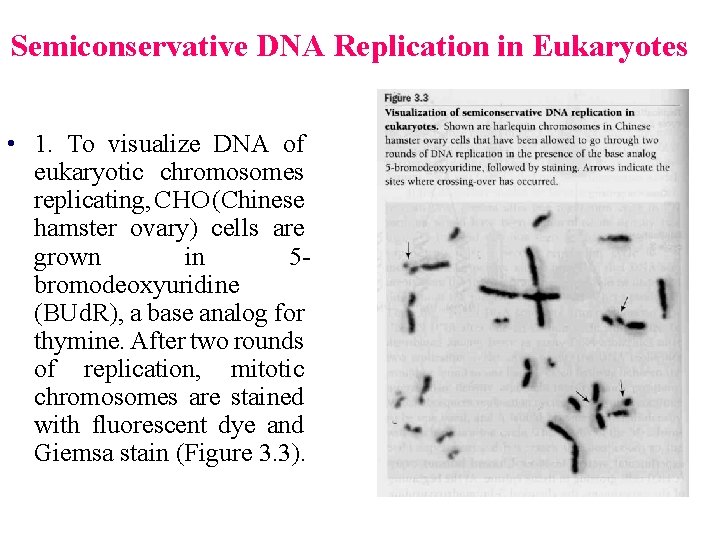
Semiconservative DNA Replication in Eukaryotes • 1. To visualize DNA of eukaryotic chromosomes replicating, CHO (Chinese hamster ovary) cells are grown in 5 bromodeoxyuridine (BUd. R), a base analog for thymine. After two rounds of replication, mitotic chromosomes are stained with fluorescent dye and Giemsa stain (Figure 3. 3).

• 2. DNA containing T stains darkly, while DNA containing two BUd. R strands stains lightly. Observed that after one generation, both chromatids stain the same, each with one BUd. R strand one T strand. After two generations, they stain differently and are called harlequin chromosomes, one light (both strands have BUd. R) and one dark (one strand has BUd. R and other strand has T). • 3. Showed that eukaryotes also use semiconservative DNA replication.

Enzymes Involved in DNA Synthesis • 1. First isolation of an enzyme involved in DNA replication was in 1955. Arthur Kornberg won the 1959 Nobel Prize in Physiology or Medicine for this work.

DNA Polymerase I • 1. Accomplished in vitro synthesis of E. coli DNA. His reaction mixture included: • a. DNA fragments (template). • b. Radioactively labeled d. NTPs (d. ATP, d. GTP, d. TTP and d. CTP). • c. E. coli lysate. • 2. Enzyme originally called the Kornberg enzyme, now known as DNA Polymerase I. Once isolated, could characterize its activity, showing that the above components are required, along with Mg 2+ ions for maximum activity.

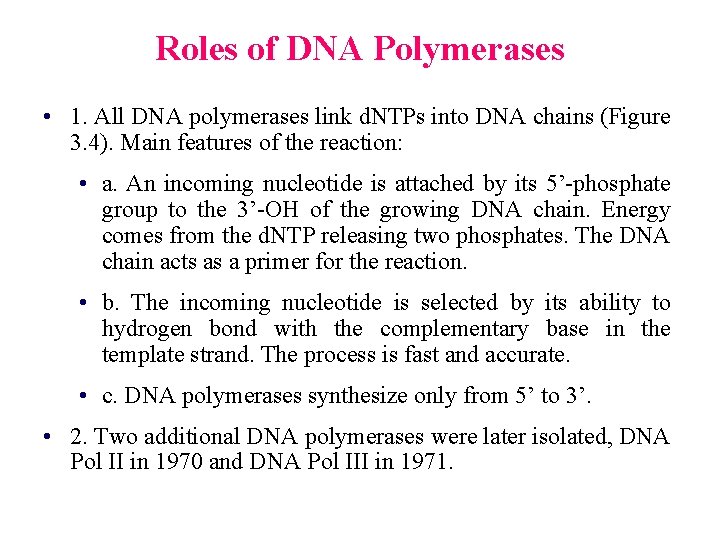
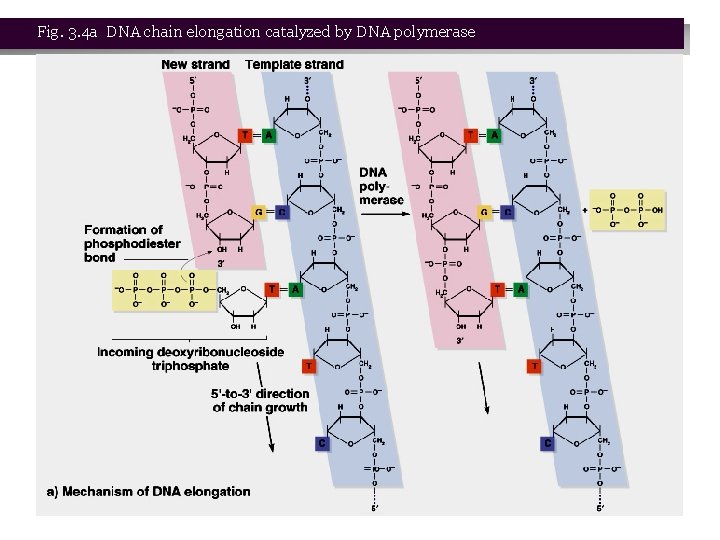
Roles of DNA Polymerases • 1. All DNA polymerases link d. NTPs into DNA chains (Figure 3. 4). Main features of the reaction: • a. An incoming nucleotide is attached by its 5’-phosphate group to the 3’-OH of the growing DNA chain. Energy comes from the d. NTP releasing two phosphates. The DNA chain acts as a primer for the reaction. • b. The incoming nucleotide is selected by its ability to hydrogen bond with the complementary base in the template strand. The process is fast and accurate. • c. DNA polymerases synthesize only from 5’ to 3’. • 2. Two additional DNA polymerases were later isolated, DNA Pol II in 1970 and DNA Pol III in 1971.

Fig. 3. 4 a DNA chain elongation catalyzed by DNA polymerase

Fig. 3. 4 b DNA chain elongation catalyzed by DNA polymerase Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

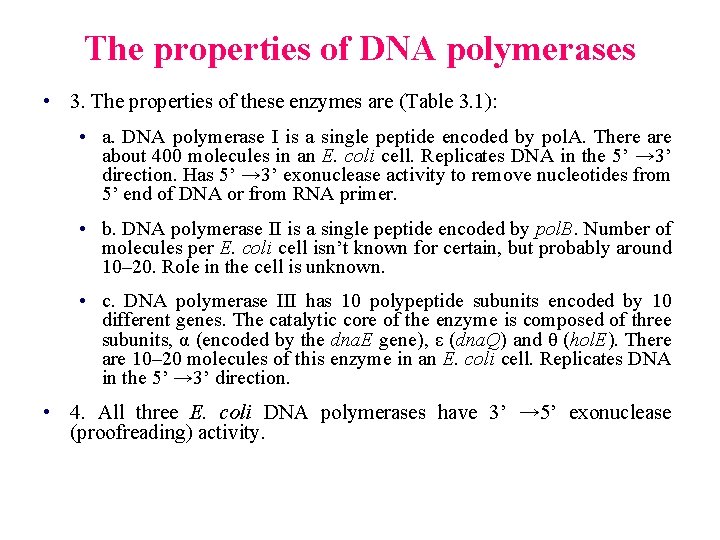
The properties of DNA polymerases • 3. The properties of these enzymes are (Table 3. 1): • a. DNA polymerase I is a single peptide encoded by pol. A. There about 400 molecules in an E. coli cell. Replicates DNA in the 5’ → 3’ direction. Has 5’ → 3’ exonuclease activity to remove nucleotides from 5’ end of DNA or from RNA primer. • b. DNA polymerase II is a single peptide encoded by pol. B. Number of molecules per E. coli cell isn’t known for certain, but probably around 10– 20. Role in the cell is unknown. • c. DNA polymerase III has 10 polypeptide subunits encoded by 10 different genes. The catalytic core of the enzyme is composed of three subunits, α (encoded by the dna. E gene), ε (dna. Q) and θ (hol. E). There are 10– 20 molecules of this enzyme in an E. coli cell. Replicates DNA in the 5’ → 3’ direction. • 4. All three E. coli DNA polymerases have 3’ → 5’ exonuclease (proofreading) activity.

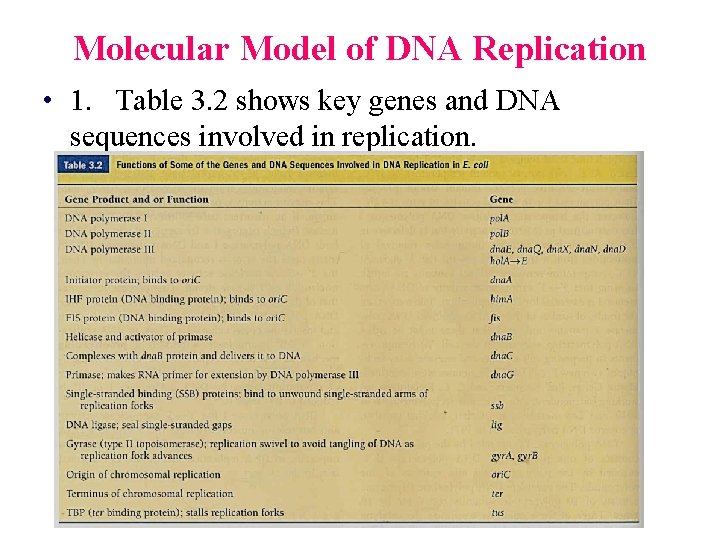
Molecular Model of DNA Replication • 1. Table 3. 2 shows key genes and DNA sequences involved in replication.

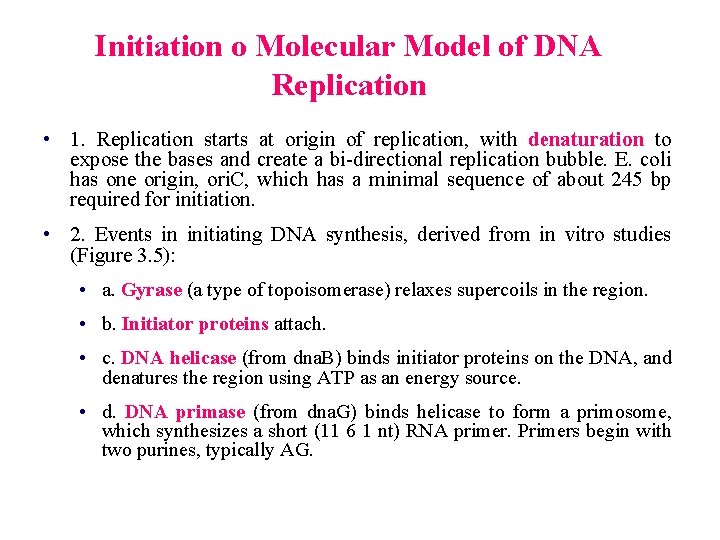
Initiation o Molecular Model of DNA Replication • 1. Replication starts at origin of replication, with denaturation to expose the bases and create a bi-directional replication bubble. E. coli has one origin, ori. C, which has a minimal sequence of about 245 bp required for initiation. • 2. Events in initiating DNA synthesis, derived from in vitro studies (Figure 3. 5): • a. Gyrase (a type of topoisomerase) relaxes supercoils in the region. • b. Initiator proteins attach. • c. DNA helicase (from dna. B) binds initiator proteins on the DNA, and denatures the region using ATP as an energy source. • d. DNA primase (from dna. G) binds helicase to form a primosome, which synthesizes a short (11 6 1 nt) RNA primer. Primers begin with two purines, typically AG.

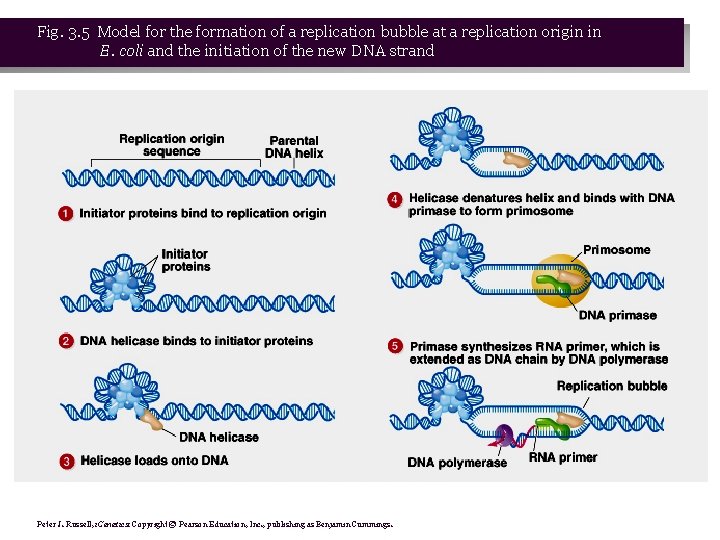
Fig. 3. 5 Model for the formation of a replication bubble at a replication origin in E. coli and the initiation of the new DNA strand Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

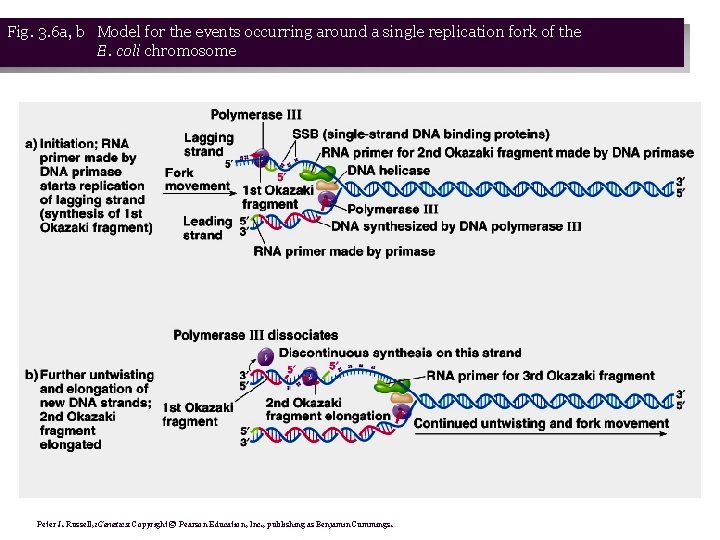
Fig. 3. 6 a, b Model for the events occurring around a single replication fork of the E. coli chromosome Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

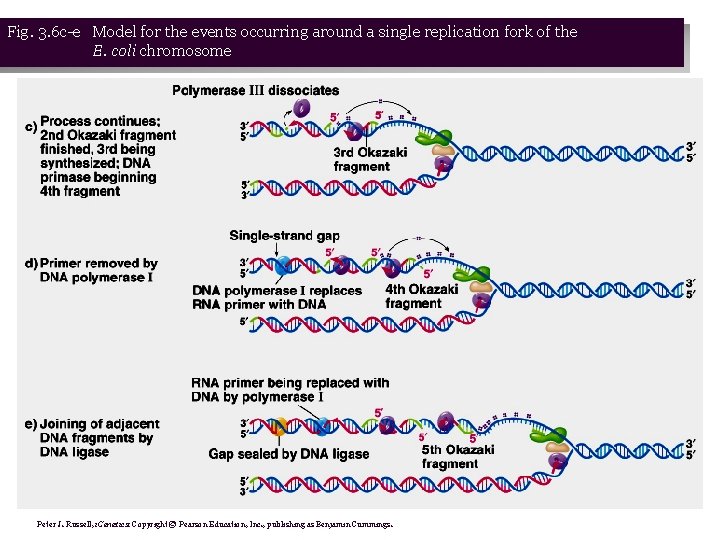
Fig. 3. 6 c-e Model for the events occurring around a single replication fork of the E. coli chromosome Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

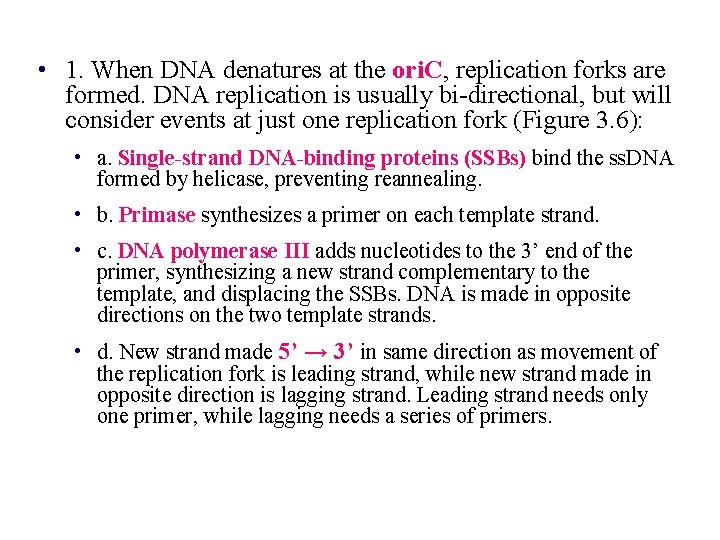
• 1. When DNA denatures at the ori. C, replication forks are formed. DNA replication is usually bi-directional, but will consider events at just one replication fork (Figure 3. 6): • a. Single-strand DNA-binding proteins (SSBs) bind the ss. DNA formed by helicase, preventing reannealing. • b. Primase synthesizes a primer on each template strand. • c. DNA polymerase III adds nucleotides to the 3’ end of the primer, synthesizing a new strand complementary to the template, and displacing the SSBs. DNA is made in opposite directions on the two template strands. • d. New strand made 5’ → 3’ in same direction as movement of the replication fork is leading strand, while new strand made in opposite direction is lagging strand. Leading strand needs only one primer, while lagging needs a series of primers.

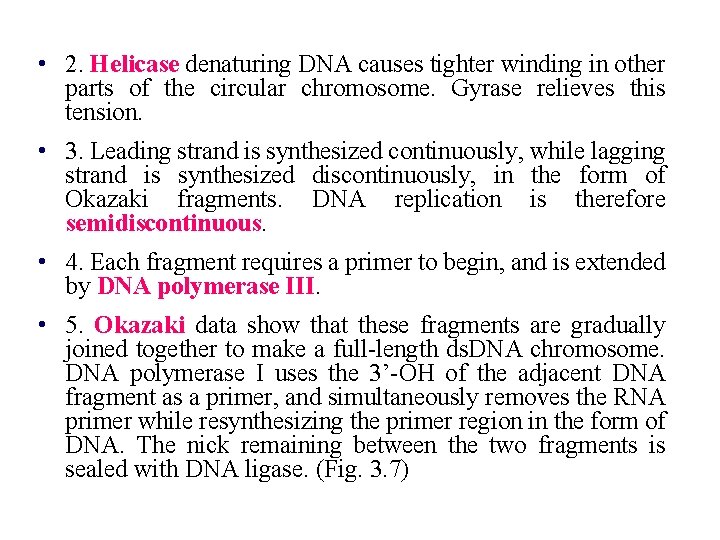
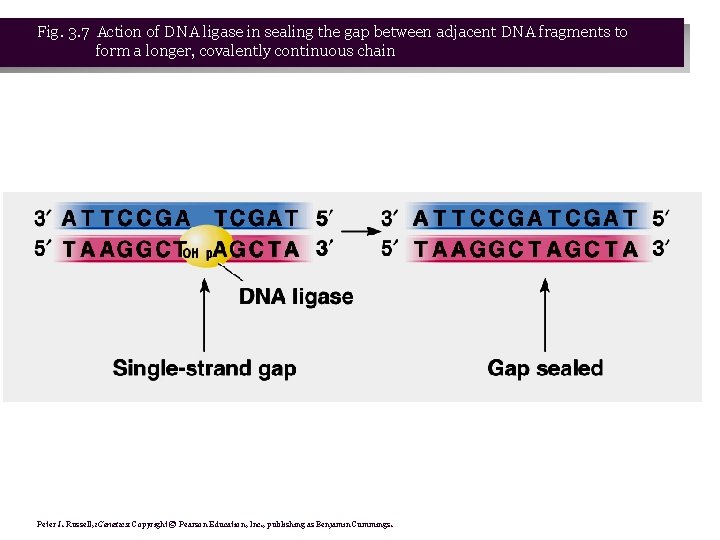
• 2. Helicase denaturing DNA causes tighter winding in other parts of the circular chromosome. Gyrase relieves this tension. • 3. Leading strand is synthesized continuously, while lagging strand is synthesized discontinuously, in the form of Okazaki fragments. DNA replication is therefore semidiscontinuous. • 4. Each fragment requires a primer to begin, and is extended by DNA polymerase III. • 5. Okazaki data show that these fragments are gradually joined together to make a full-length ds. DNA chromosome. DNA polymerase I uses the 3’-OH of the adjacent DNA fragment as a primer, and simultaneously removes the RNA primer while resynthesizing the primer region in the form of DNA. The nick remaining between the two fragments is sealed with DNA ligase. (Fig. 3. 7)

Fig. 3. 7 Action of DNA ligase in sealing the gap between adjacent DNA fragments to form a longer, covalently continuous chain Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

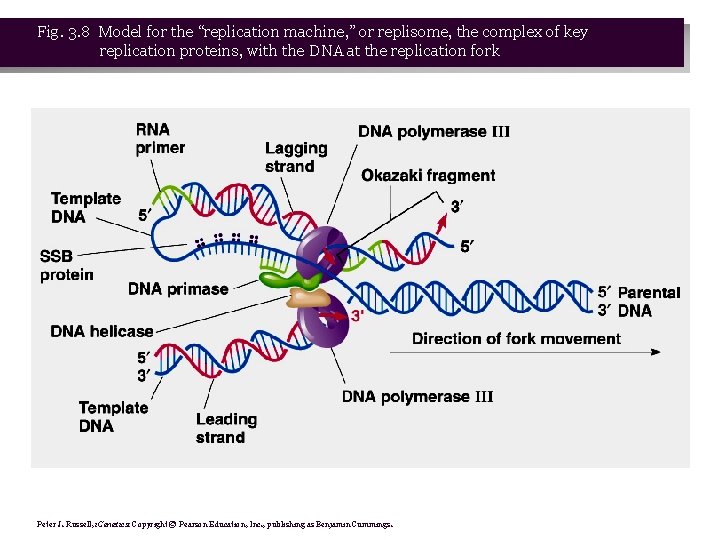
• 6. Key proteins are associated to form a replisome. Template DNA probably bends to allow synthesis of both leading and lagging strands at the replication fork (Fig. 3. 8) • 7. Early stages of bidirectional replication are summarized in Figure 3. 9.

Fig. 3. 8 Model for the “replication machine, ” or replisome, the complex of key replication proteins, with the DNA at the replication fork Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.