Chapter 9 DNA Separation Detection Fundamentals of Forensic



- Slides: 26

Chapter 9 DNA Separation & Detection Fundamentals of Forensic DNA Typing Slides prepared by John M. Butler June 2009

Chapter 9 – DNA Separations and Fluorescence Detection Chapter Summary • A multiplex PCR amplification of STR markers produces a complex mixture of DNA molecules that must be separated based on DNA size and fluorescent dye label to produce a coherent DNA profile. Original gel electrophoresis separation methods have been almost entirely replaced by capillary electrophoresis (CE) instruments over the past decade due to ease of use and automation. The most commonly used CE systems are the single capillary ABI Prism 310 Genetic Analyzer and the multicapillary ABI 3100 or 3130 xl. These CE instruments electrokinetically inject the negatively charged DNA molecules from a formamide-diluted sample of the PCR products mixed with an internal size standard. The size standard is labeled with a separate fluorescent dye to enable calibration of each analysis so that comparisons can be made between samples run at different times on the same instrument. A polymer solution inside the capillary permits resolution of DNA fragments differing by as little as a single basepair (bp) over a size range of approximately 100 to 400 bp. Fluorescent dyes are present on one strand of each PCR product due to incorporation of a PCR primer during multiplex PCR amplification. These dyes are excited by laser as they pass a detection point in the CE instrument. Since the four or five fluorescent dyes used in STR analysis have different chemical properties, they emit light at slightly different wavelengths enabling detection in different color channels. Because there is overlap with the emitted light from the different dyes, mathematical algorithms are used to perform a “matrix correction” or “spectral calibration” so that individual DNA peaks in an electropherogram appear to be labeled with a single color.

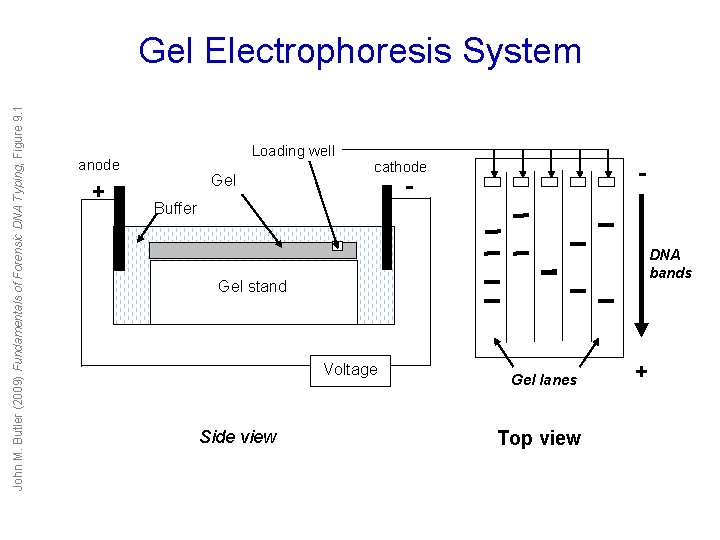
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 1 Gel Electrophoresis System Loading well anode + Gel cathode - - Buffer DNA bands Gel stand Voltage Side view Gel lanes Top view +

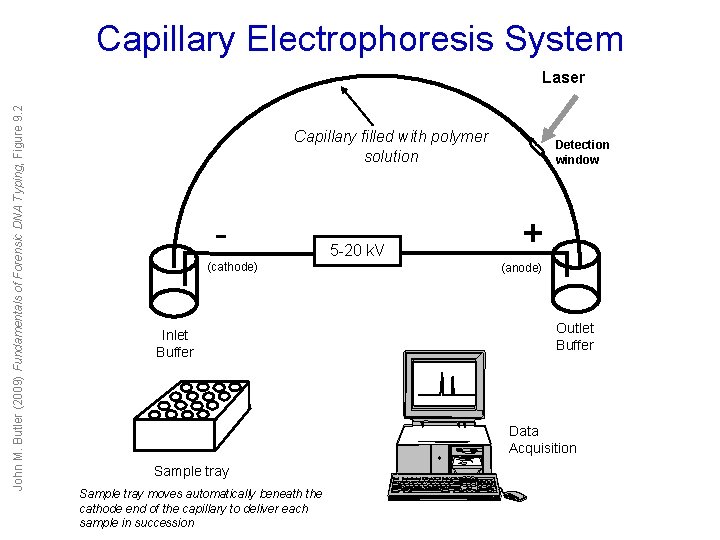
Capillary Electrophoresis System John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 2 Laser Capillary filled with polymer solution (cathode) Inlet Buffer 5 -20 k. V Detection window + (anode) Outlet Buffer Data Acquisition Sample tray moves automatically beneath the cathode end of the capillary to deliver each sample in succession

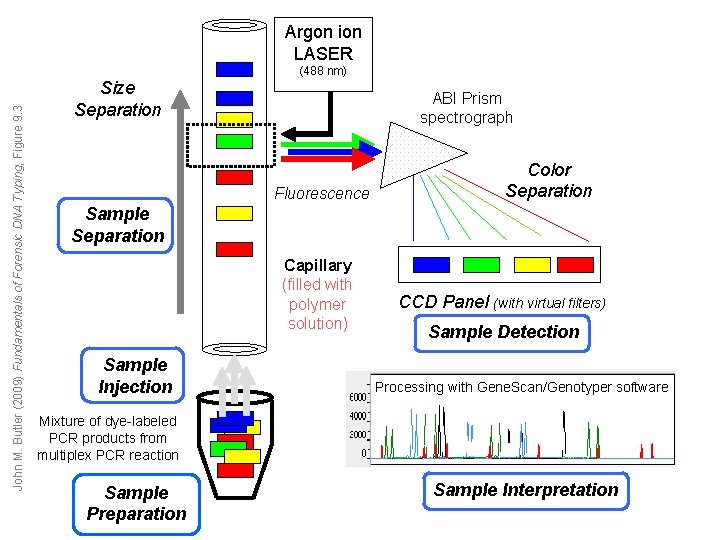
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 3 Argon ion LASER Size Separation (488 nm) ABI Prism spectrograph Fluorescence Color Separation Sample Separation Capillary (filled with polymer solution) Sample Injection CCD Panel (with virtual filters) Sample Detection Processing with Gene. Scan/Genotyper software Mixture of dye-labeled PCR products from multiplex PCR reaction Sample Preparation Sample Interpretation

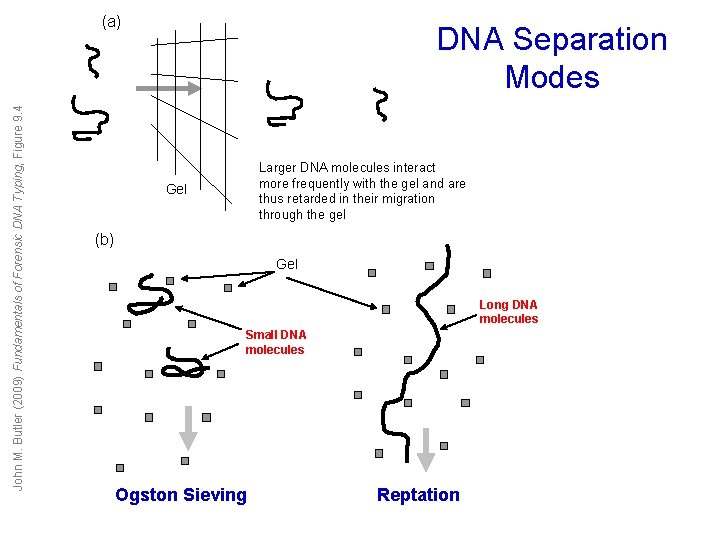
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 4 (a) DNA Separation Modes Larger DNA molecules interact more frequently with the gel and are thus retarded in their migration through the gel Gel (b) Gel Long DNA molecules Small DNA molecules Ogston Sieving Reptation

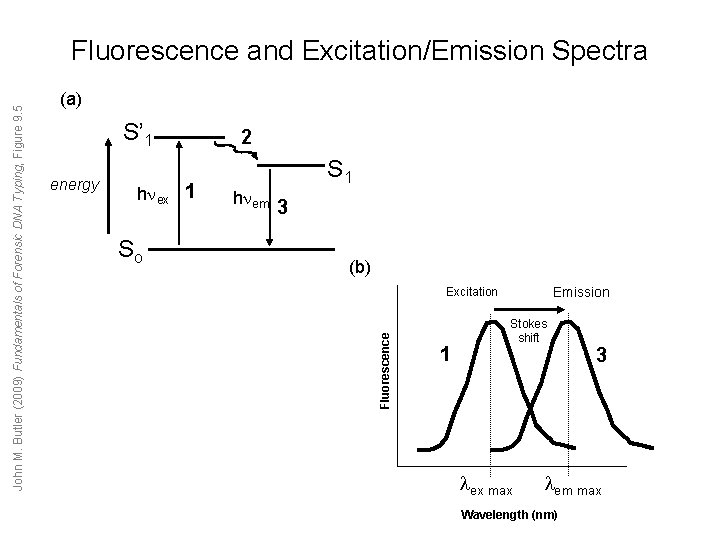
(a) S’ 1 energy h ex 1 So 2 S 1 h em 3 (b) Emission Excitation Fluorescence John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 5 Fluorescence and Excitation/Emission Spectra 1 Stokes shift ex max 3 em max Wavelength (nm)

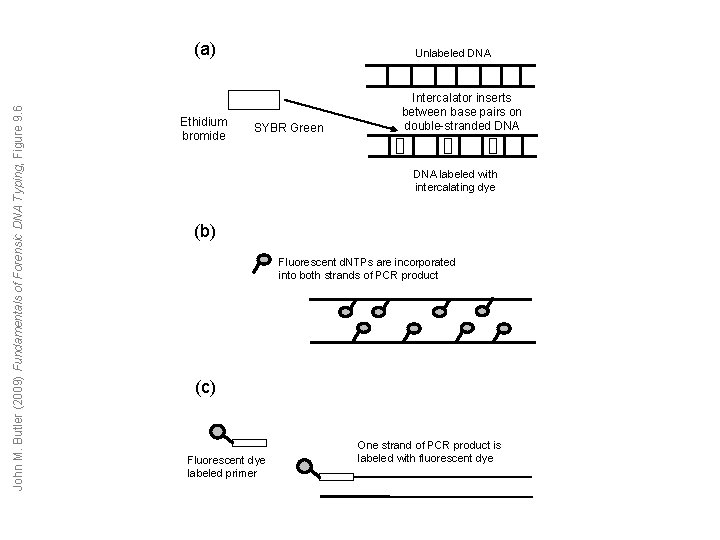
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 6 (a) Ethidium bromide Unlabeled DNA SYBR Green Intercalator inserts between base pairs on double-stranded DNA labeled with intercalating dye (b) Fluorescent d. NTPs are incorporated into both strands of PCR product (c) Fluorescent dye labeled primer One strand of PCR product is labeled with fluorescent dye

John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 7 FAM (blue) JOE (green) TAMRA ROX (yellow) (red)

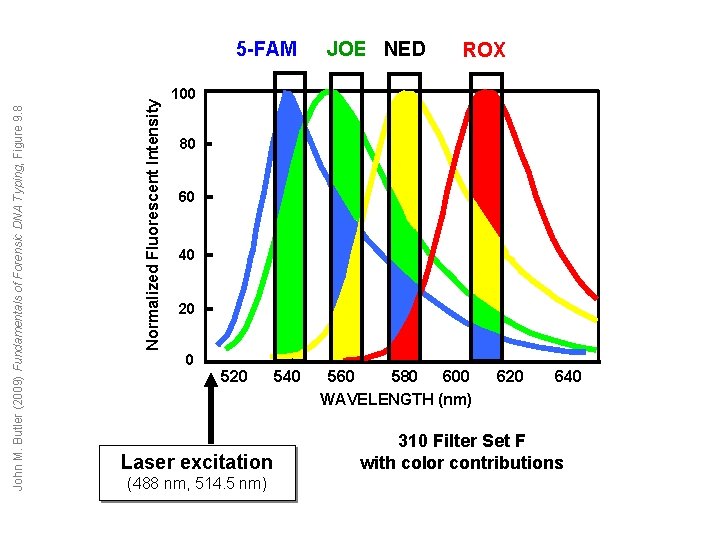
Normalized Fluorescent Intensity John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 8 5 -FAM JOE NED ROX 100 80 60 40 20 0 520 Laser excitation (488 nm, 514. 5 nm) 540 600 560 580 WAVELENGTH (nm) 620 640 310 Filter Set F with color contributions

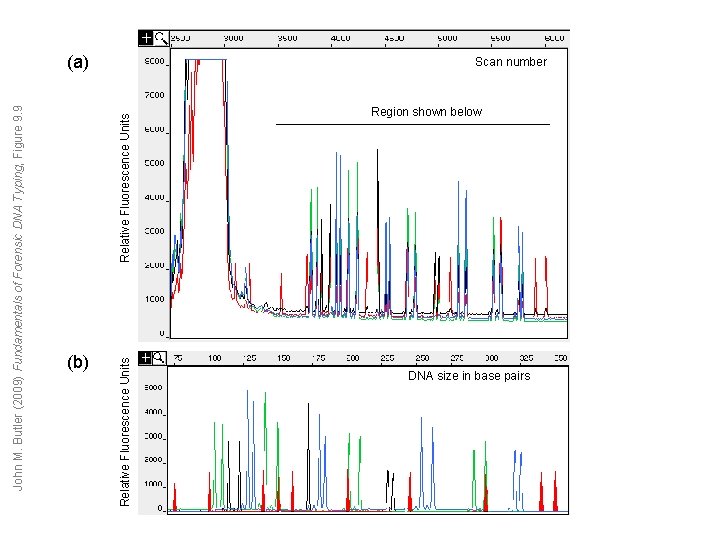
(b) Relative Fluorescence Units John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 9 (a) Scan number Region shown below DNA size in base pairs

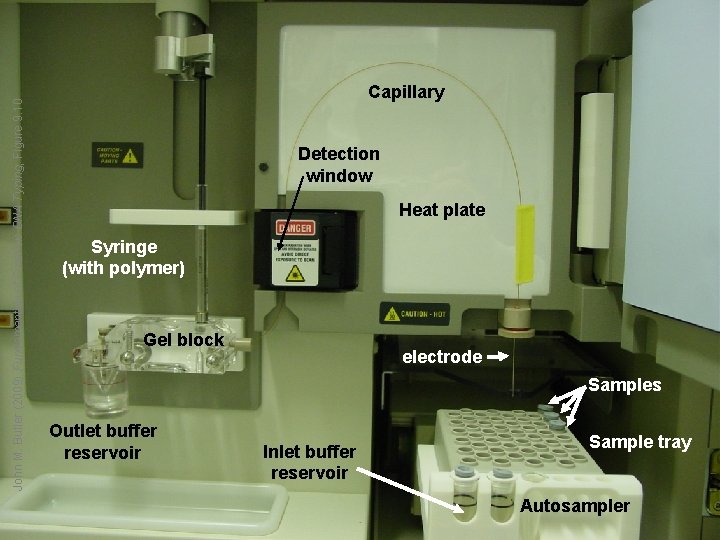
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 10 Capillary Detection window Heat plate Syringe (with polymer) Gel block electrode Samples Outlet buffer reservoir Inlet buffer reservoir Sample tray Autosampler

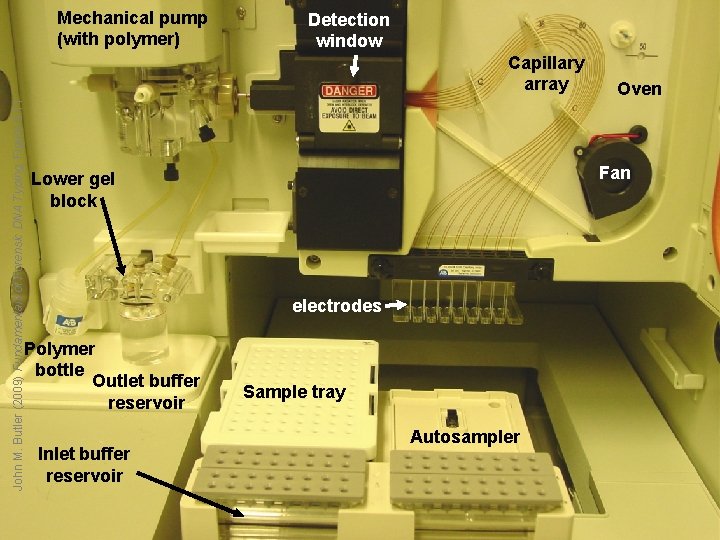
Mechanical pump (with polymer) Detection window John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 11 Capillary array Fan Lower gel block electrodes Polymer bottle Outlet buffer reservoir Inlet buffer reservoir Oven Sample tray Autosampler

John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 9. 12 Capillaries Electrodes for Injection

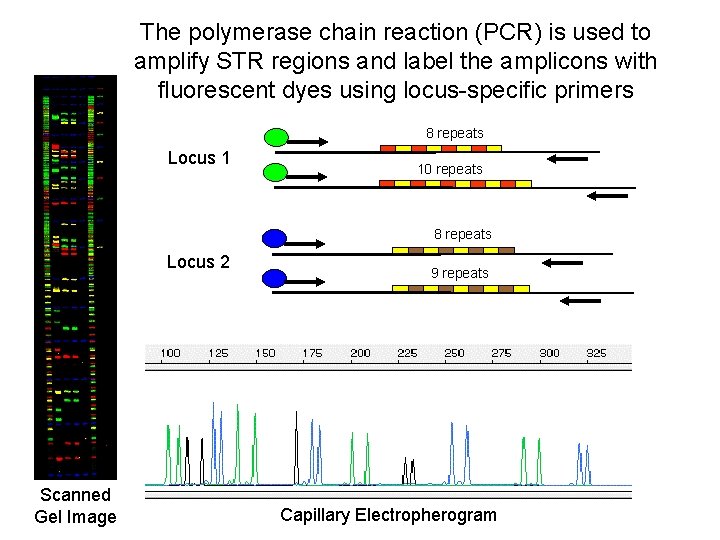
The polymerase chain reaction (PCR) is used to amplify STR regions and label the amplicons with fluorescent dyes using locus-specific primers 8 repeats Locus 1 10 repeats 8 repeats Locus 2 Scanned Gel Image 9 repeats Capillary Electropherogram

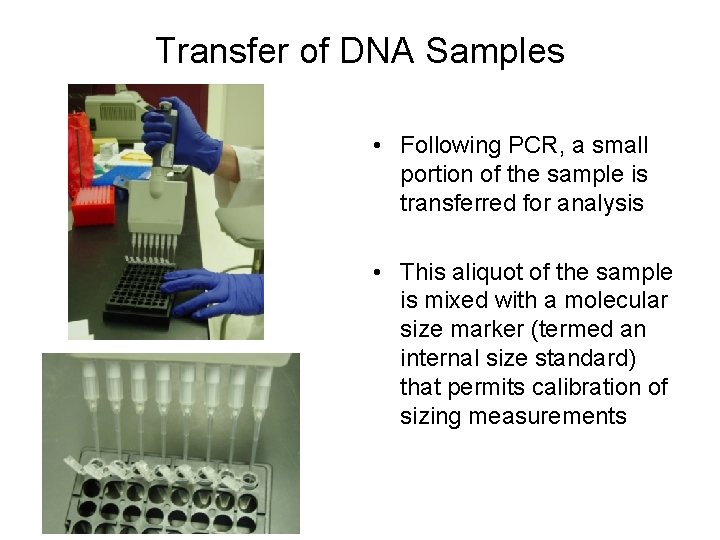
Transfer of DNA Samples • Following PCR, a small portion of the sample is transferred for analysis • This aliquot of the sample is mixed with a molecular size marker (termed an internal size standard) that permits calibration of sizing measurements

Sample Plates Spun Down via a Centrifuge • Sample plates are spun to remove bubbles that would interfere with the injection (loading) process onto the capillary electrophoresis instrument

ABI 3130 xl DNA Analysis Instrument • Import sample names • Determine run conditions (voltages and times to be used based on laboratory protocols)

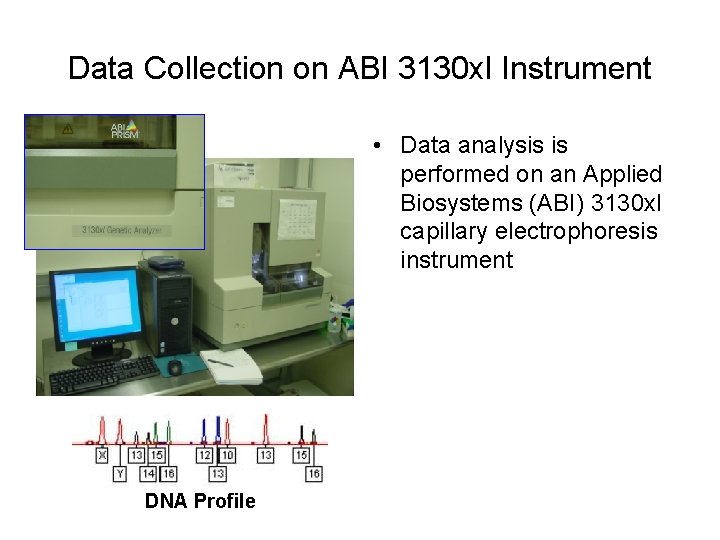
Data Collection on ABI 3130 xl Instrument • Data analysis is performed on an Applied Biosystems (ABI) 3130 xl capillary electrophoresis instrument DNA Profile

Capillary Electrophoresis Instrumentation ABI 310 single capillary ABI 3100 16 capillary array

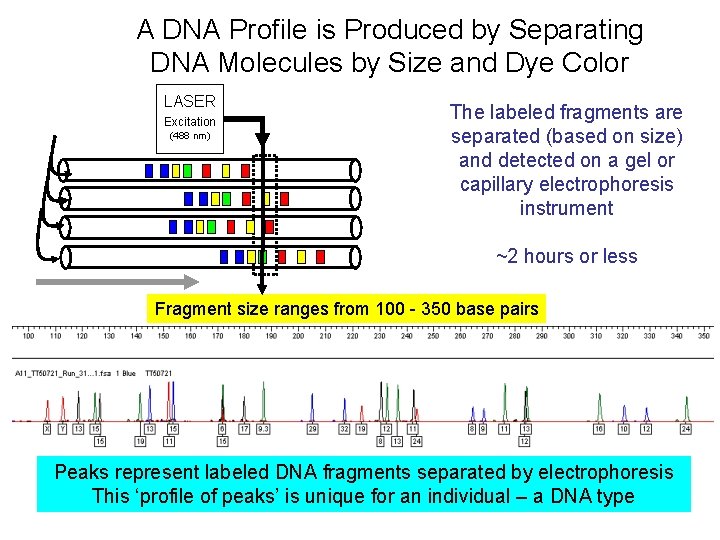
A DNA Profile is Produced by Separating DNA Molecules by Size and Dye Color LASER Excitation (488 nm) The labeled fragments are separated (based on size) and detected on a gel or capillary electrophoresis instrument ~2 hours or less Fragment size ranges from 100 - 350 base pairs Peaks represent labeled DNA fragments separated by electrophoresis This ‘profile of peaks’ is unique for an individual – a DNA type

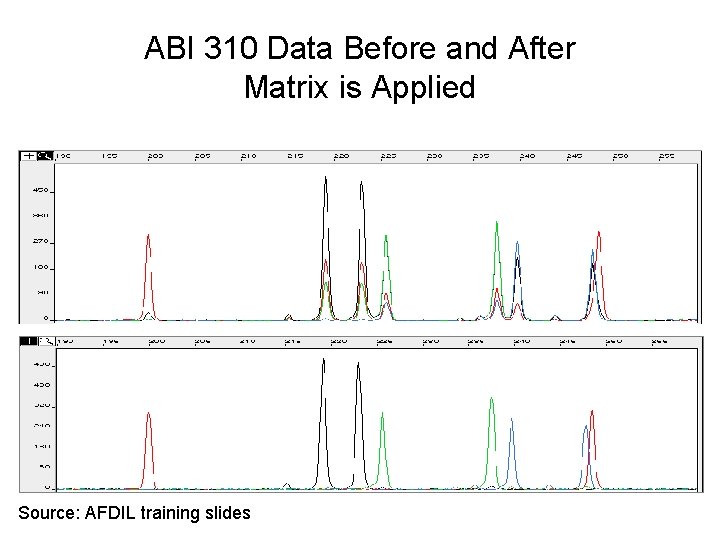
ABI 310 Data Before and After Matrix is Applied Source: AFDIL training slides

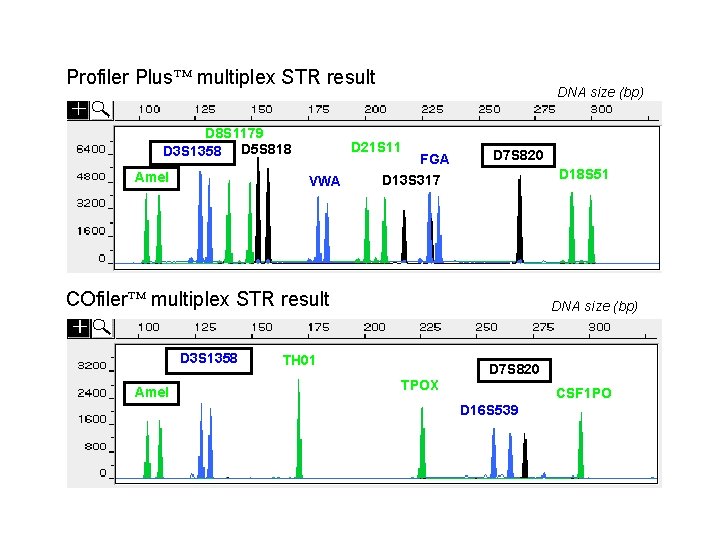
Profiler Plus multiplex STR result D 8 S 1179 D 3 S 1358 D 5 S 818 Amel DNA size (bp) D 21 S 11 VWA FGA D 7 S 820 D 18 S 51 D 13 S 317 COfiler multiplex STR result D 3 S 1358 Amel DNA size (bp) TH 01 D 7 S 820 TPOX CSF 1 PO D 16 S 539

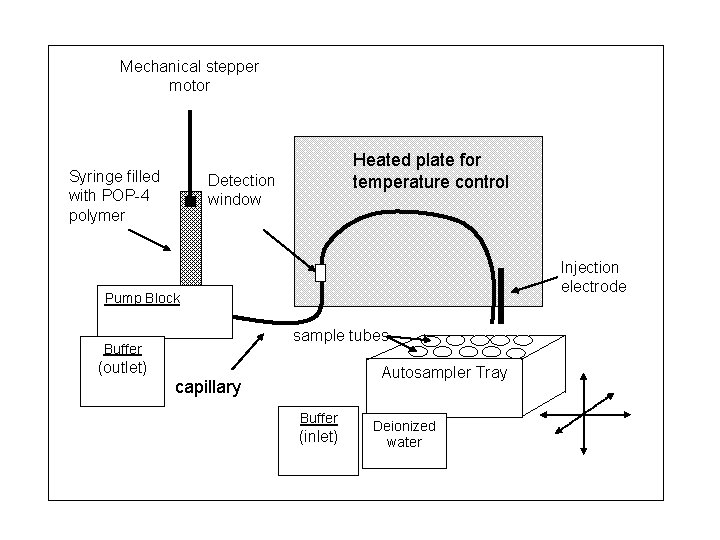
Mechanical stepper motor Syringe filled with POP-4 polymer Heated plate for temperature control Detection window Injection electrode Pump Block sample tubes Buffer (outlet) Autosampler Tray capillary Buffer (inlet) Deionized water

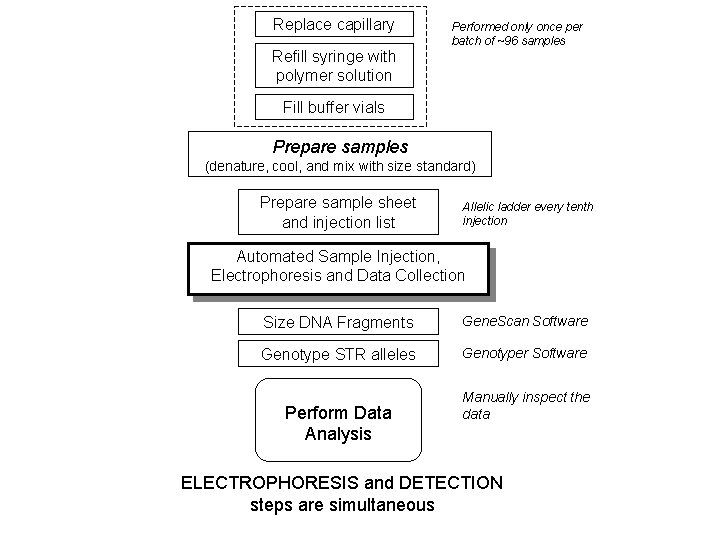
Replace capillary Performed only once per batch of ~96 samples Refill syringe with polymer solution Fill buffer vials Prepare samples (denature, cool, and mix with size standard) Prepare sample sheet and injection list Allelic ladder every tenth injection Automated Sample Injection, Electrophoresis and Data Collection Size DNA Fragments Gene. Scan Software Genotype STR alleles Genotyper Software Perform Data Analysis Manually inspect the data ELECTROPHORESIS and DETECTION steps are simultaneous

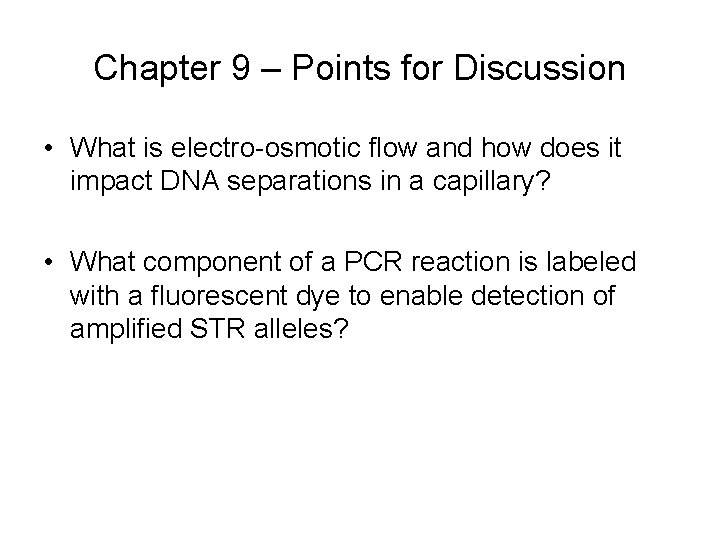
Chapter 9 – Points for Discussion • What is electro-osmotic flow and how does it impact DNA separations in a capillary? • What component of a PCR reaction is labeled with a fluorescent dye to enable detection of amplified STR alleles?