Chapter 8 STR Markers Fundamentals of Forensic DNA







- Slides: 36

Chapter 8 STR Markers Fundamentals of Forensic DNA Typing Slides prepared by John M. Butler June 2009

Chapter 8 – STR Markers Chapter Summary Short tandem repeats (STRs) are accordion-like regions of the human genome that vary in length between people based on a repeated DNA sequence. Forensic laboratories commonly use tetranucleotide repeats, containing a four basepair repeat structure such as GATA, because these STR loci produce fewer artifactual stutter products than dinucleotide like CA repeats used in other applications such as genetic mapping studies. In 1997, the FBI Laboratory selected 13 STR loci that form the backbone of the U. S. national DNA database. Many of these same STR loci are used by other countries around the world. Commercial STR kits enable consistency in marker use and allele nomenclature between laboratories and off-load quality control concerns from busy forensic laboratories. STR kits are available that permit simultaneous PCR amplification of 15 STR loci. A DNA sequence found on both the X and Y chromosomes named amelogenin is commonly used for sextyping in combination with the STRs present in commercial kits.

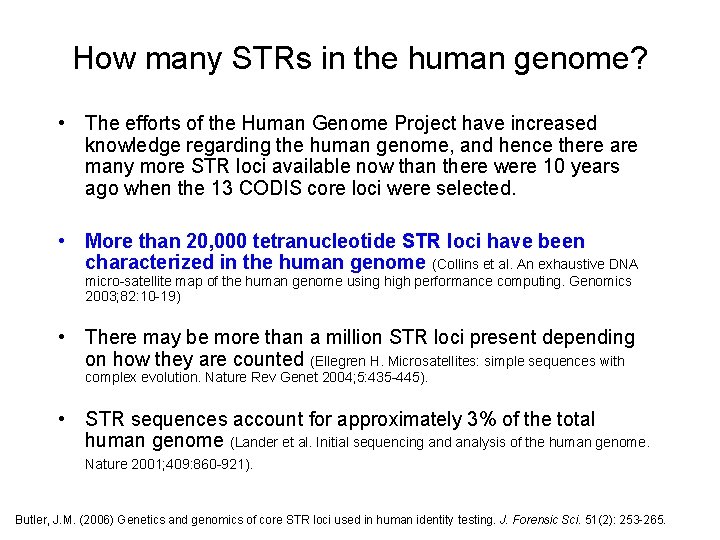
How many STRs in the human genome? • The efforts of the Human Genome Project have increased knowledge regarding the human genome, and hence there are many more STR loci available now than there were 10 years ago when the 13 CODIS core loci were selected. • More than 20, 000 tetranucleotide STR loci have been characterized in the human genome (Collins et al. An exhaustive DNA micro-satellite map of the human genome using high performance computing. Genomics 2003; 82: 10 -19) • There may be more than a million STR loci present depending on how they are counted (Ellegren H. Microsatellites: simple sequences with complex evolution. Nature Rev Genet 2004; 5: 435 -445). • STR sequences account for approximately 3% of the total human genome (Lander et al. Initial sequencing and analysis of the human genome. Nature 2001; 409: 860 -921). Butler, J. M. (2006) Genetics and genomics of core STR loci used in human identity testing. J. Forensic Sci. 51(2): 253 -265.

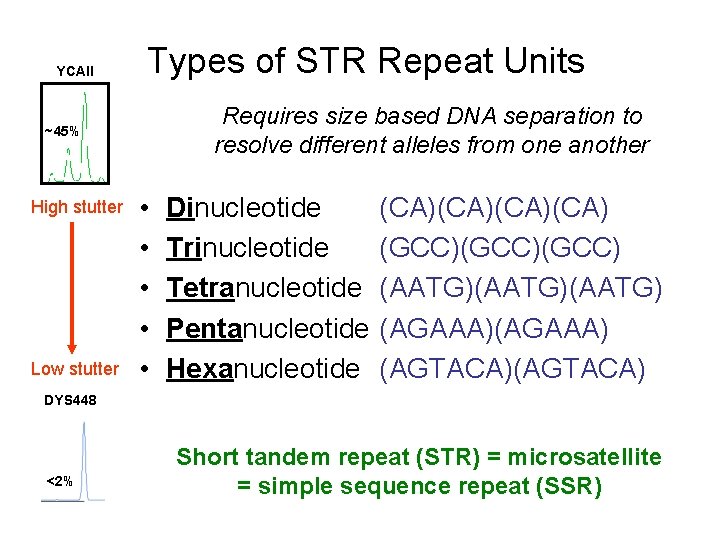
YCAII Types of STR Repeat Units Requires size based DNA separation to resolve different alleles from one another ~45% High stutter Low stutter • • • Dinucleotide Trinucleotide Tetranucleotide Pentanucleotide Hexanucleotide (CA)(CA) (GCC)(GCC) (AATG)(AATG) (AGAAA) (AGTACA) DYS 448 <2% Short tandem repeat (STR) = microsatellite = simple sequence repeat (SSR)

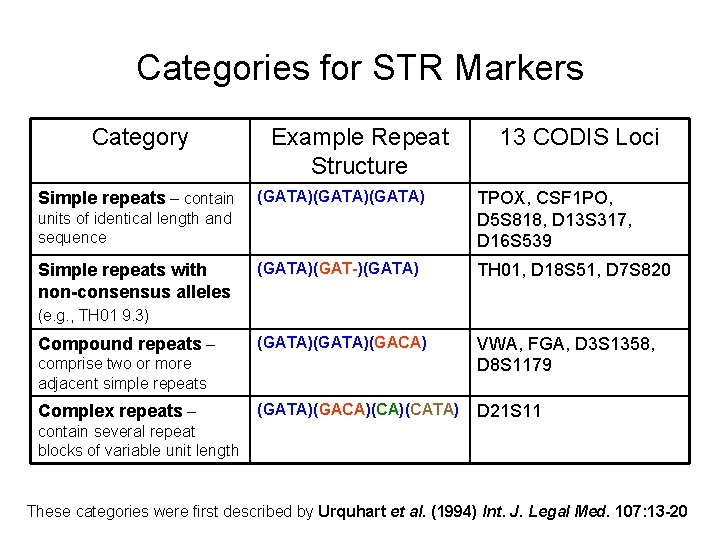
Categories for STR Markers Category Simple repeats – contain Example Repeat Structure (GATA)(GATA) TPOX, CSF 1 PO, D 5 S 818, D 13 S 317, D 16 S 539 (GATA)(GAT-)(GATA) TH 01, D 18 S 51, D 7 S 820 (GATA)(GACA) VWA, FGA, D 3 S 1358, D 8 S 1179 (GATA)(GACA)(CATA) D 21 S 11 units of identical length and sequence Simple repeats with non-consensus alleles 13 CODIS Loci (e. g. , TH 01 9. 3) Compound repeats – comprise two or more adjacent simple repeats Complex repeats – contain several repeat blocks of variable unit length These categories were first described by Urquhart et al. (1994) Int. J. Legal Med. 107: 13 -20

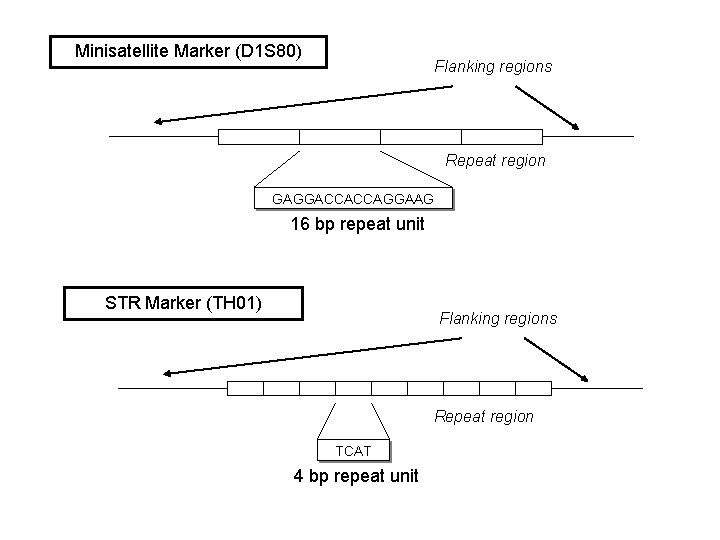
Minisatellite Marker (D 1 S 80) Flanking regions Repeat region GAGGACCACCAGGAAG 16 bp repeat unit STR Marker (TH 01) Flanking regions Repeat region TCAT 4 bp repeat unit

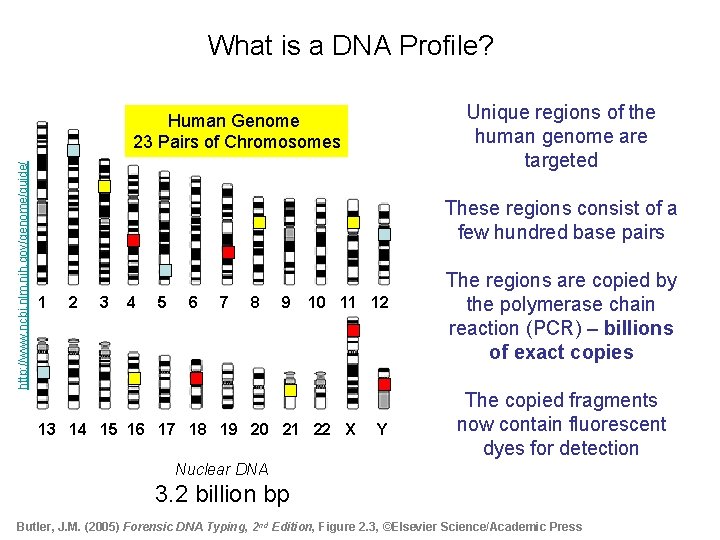
What is a DNA Profile? Unique regions of the human genome are targeted http: //www. ncbi. nlm. nih. gov/genome/guide/ Human Genome 23 Pairs of Chromosomes These regions consist of a few hundred base pairs 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 X Y The regions are copied by the polymerase chain reaction (PCR) – billions of exact copies The copied fragments now contain fluorescent dyes for detection Nuclear DNA 3. 2 billion bp Butler, J. M. (2005) Forensic DNA Typing, 2 nd Edition, Figure 2. 3, ©Elsevier Science/Academic Press

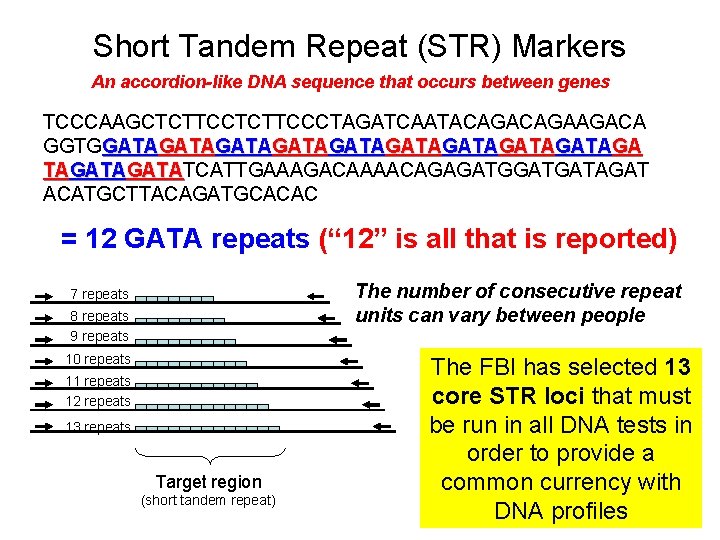
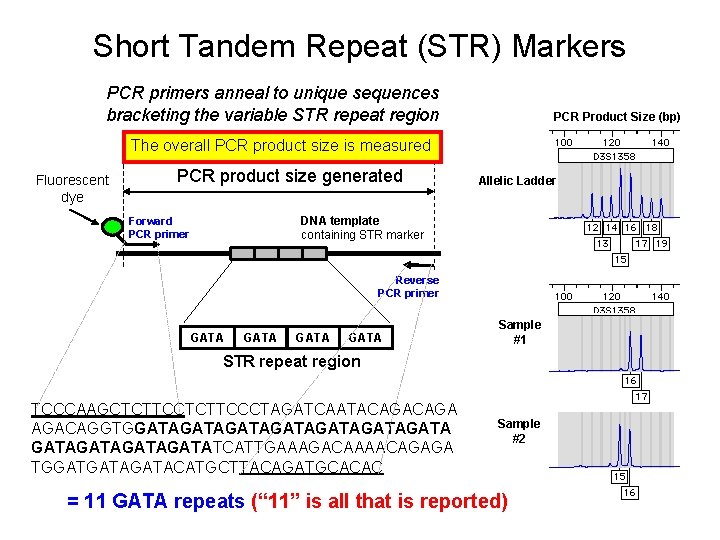
Short Tandem Repeat (STR) Markers An accordion-like DNA sequence that occurs between genes TCCCAAGCTCTTCCCTAGATCAATACAGAAGACA GGTGGATAGATAGATAGATAGATAGATATCATTGAAAGACAAAACAGAGATGATAGAT GATA ACATGCTTACAGATGCACAC = 12 GATA repeats (“ 12” is all that is reported) The number of consecutive repeat units can vary between people 7 repeats 8 repeats 9 repeats 10 repeats 11 repeats 12 repeats 13 repeats Target region (short tandem repeat) The FBI has selected 13 core STR loci that must be run in all DNA tests in order to provide a common currency with DNA profiles

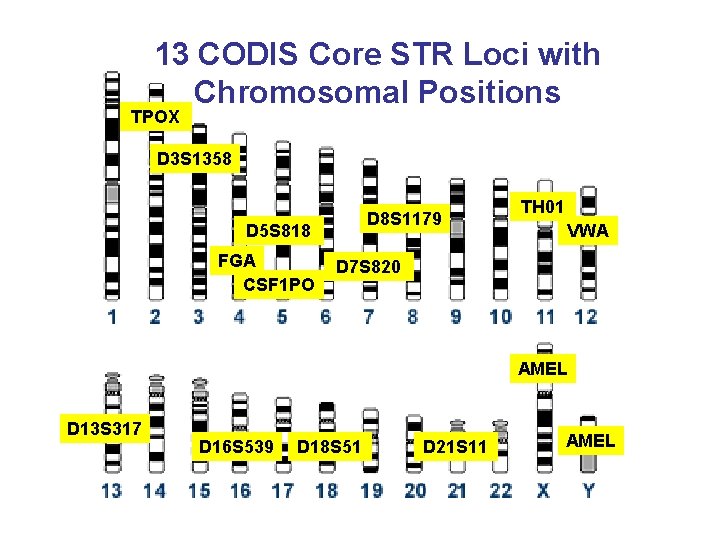
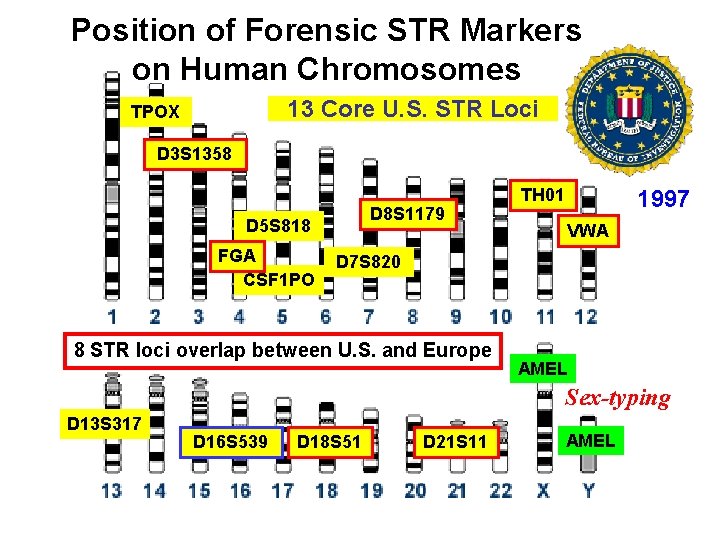
13 CODIS Core STR Loci with Chromosomal Positions TPOX D 3 S 1358 D 8 S 1179 D 5 S 818 FGA CSF 1 PO TH 01 VWA D 7 S 820 AMEL D 13 S 317 D 16 S 539 D 18 S 51 D 21 S 11 AMEL

Advantages for STR Markers • Small product sizes are generally compatible with degraded DNA and PCR enables recovery of information from small amounts of material • Multiplex amplification with fluorescence detection enables high power of discrimination in a single test • Commercially available in an easy to use kit format • Uniform set of core STR loci provide capability for national and international sharing of criminal DNA profiles

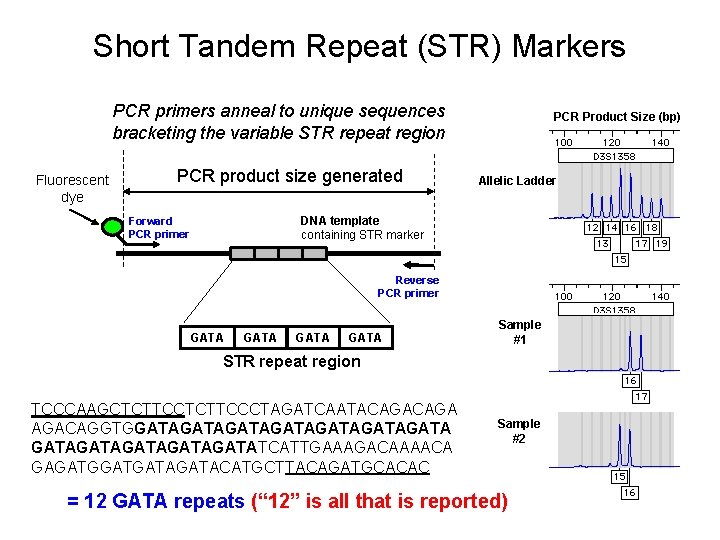
Short Tandem Repeat (STR) Markers PCR primers anneal to unique sequences bracketing the variable STR repeat region Fluorescent dye PCR product size generated PCR Product Size (bp) Allelic Ladder DNA template containing STR marker Forward PCR primer Reverse PCR primer GATA Sample #1 STR repeat region TCCCAAGCTCTTCCCTAGATCAATACAGA AGACAGGTGGATAGATAGATAGATATCATTGAAAGACAAAACA GAGATGATAGATACATGCTTACAGATGCACAC Sample #2 = 12 GATA repeats (“ 12” is all that is reported)

DNA Reaction Setup • DNA sample is added (about 1 ng based on DNA quantitation performed) – 10 µL • PCR primers and other reaction chemicals from an STR typing kit are added – 15 µL Strip of 8 tubes containing ~25 µL of solution

STR Typing Kit • Kit Components: – Primer mix – PCR Reaction Buffer and Building Blocks – DNA Polymerase (Taq Gold) • Most expensive reagent • Common kits used: – Identifiler (Applied Biosystems) – Profiler Plus/COfiler (Applied Biosystems) – Power. Plex 16 (Promega)

What is in an STR Typing Kit? • Primer mix – containing fluorescently labeled oligonucleotides used to target specific regions of the human genome – Applied Biosystems has not published their primer sequences – Power. Plex 16, which amplifies 16 genomic sites, contains 32 PCR primers

PCR Primers in an STR Kit DNA Profile

Short Tandem Repeat (STR) Markers PCR primers anneal to unique sequences bracketing the variable STR repeat region PCR Product Size (bp) The overall PCR product size is measured Fluorescent dye PCR product size generated Allelic Ladder DNA template containing STR marker Forward PCR primer Reverse PCR primer GATA Sample #1 STR repeat region TCCCAAGCTCTTCCCTAGATCAATACAGA AGACAGGTGGATAGATAGATAGATATCATTGAAAGACAAAACAGAGA TGGATGATACATGCTTACAGATGCACAC Sample #2 = 11 GATA repeats (“ 11” is all that is reported)

Position of Forensic STR Markers on Human Chromosomes 13 Core U. S. STR Loci TPOX D 3 S 1358 D 8 S 1179 D 5 S 818 FGA CSF 1 PO TH 01 1997 VWA D 7 S 820 8 STR loci overlap between U. S. and Europe AMEL Sex-typing D 13 S 317 D 16 S 539 D 18 S 51 D 21 S 11 AMEL

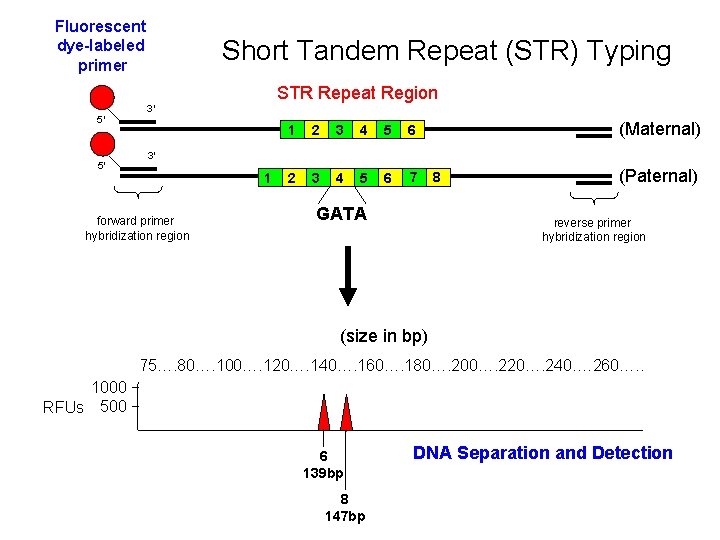
Fluorescent dye-labeled primer Short Tandem Repeat (STR) Typing STR Repeat Region 5′ 5′ 3′ 1 2 3 4 5 6 7 (Maternal) 3′ 1 forward primer hybridization region GATA 8 (Paternal) reverse primer hybridization region (size in bp) 75…. 80…. 100…. 120…. 140…. 160…. 180…. 200…. 220…. 240. … 260…. . 1000 RFUs 500 6 139 bp 8 147 bp DNA Separation and Detection

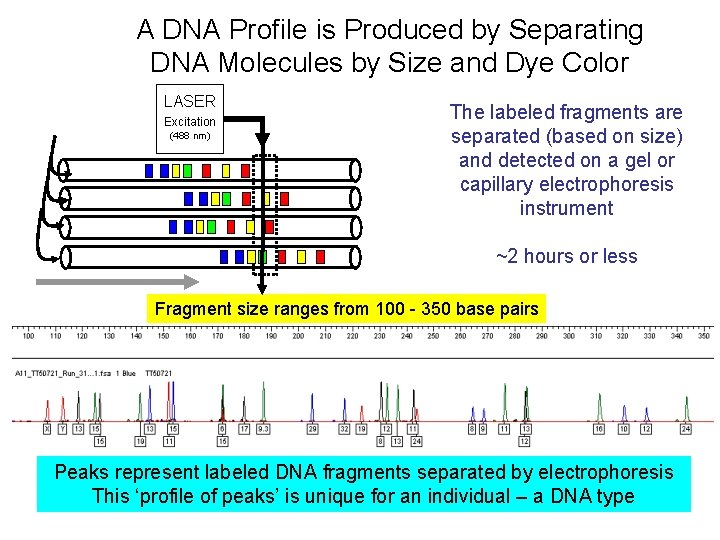
A DNA Profile is Produced by Separating DNA Molecules by Size and Dye Color LASER Excitation (488 nm) The labeled fragments are separated (based on size) and detected on a gel or capillary electrophoresis instrument ~2 hours or less Fragment size ranges from 100 - 350 base pairs Peaks represent labeled DNA fragments separated by electrophoresis This ‘profile of peaks’ is unique for an individual – a DNA type

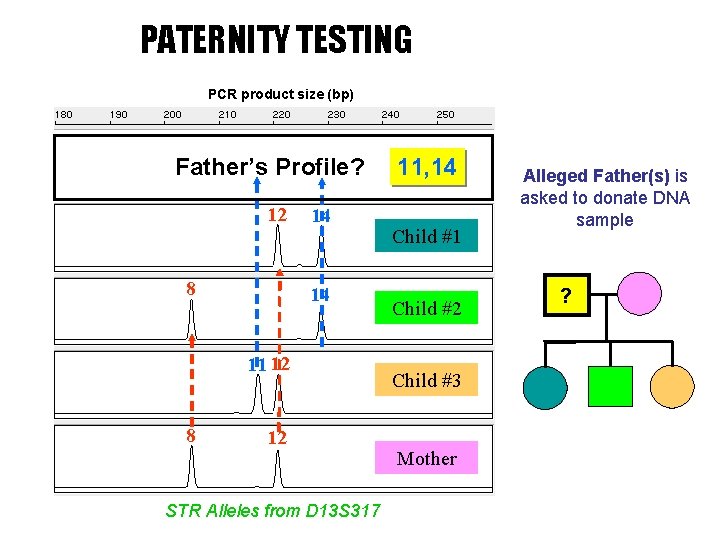
PATERNITY TESTING PCR product size (bp) 11 14 Father’s Profile? 12 8 14 14 11 12 8 12 STR Alleles from D 13 S 317 Father 11, 14 Child #1 Child #2 Child #3 Mother Alleged Father(s) is asked to donate DNA sample ?

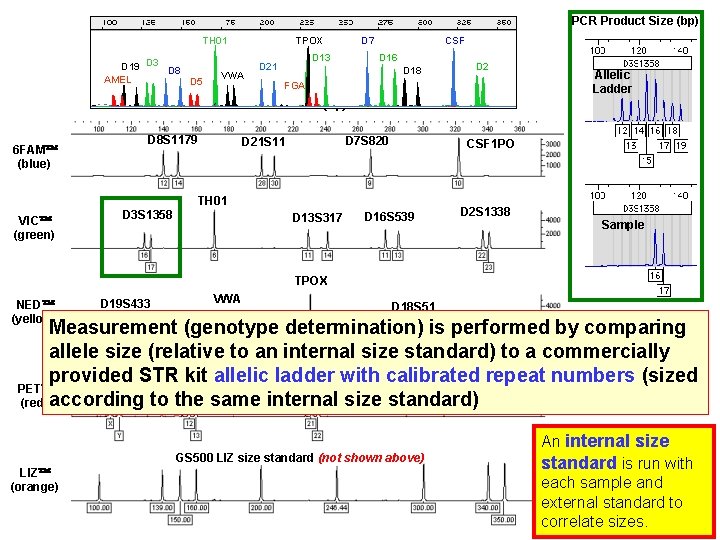
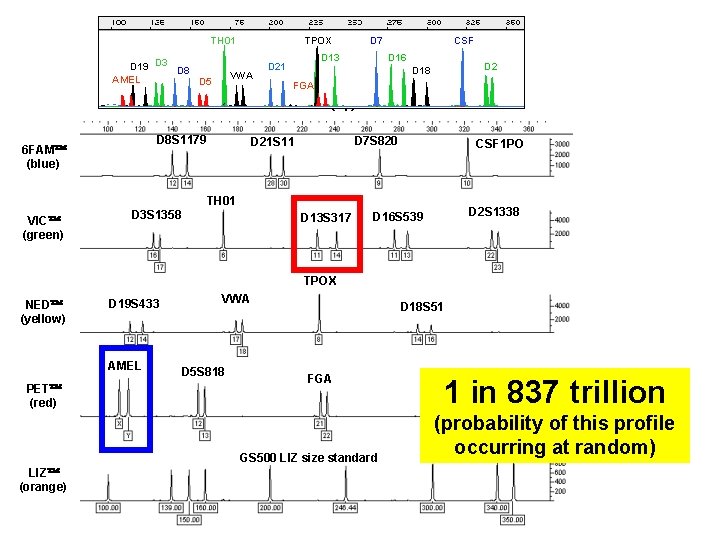
PCR Product Size (bp) TH 01 D 19 D 3 D 8 AMEL D 5 TPOX VWA D 7 D 13 D 21 CSF D 16 D 18 D 2 FGA DNA Size (bp) 6 FAM (blue) D 8 S 1179 D 7 S 820 D 21 S 11 CSF 1 PO TH 01 VIC (green) D 3 S 1358 D 13 S 317 Allelic Ladder D 16 S 539 D 2 S 1338 Sample TPOX NED (yellow) D 19 S 433 VWA D 18 S 51 Measurement (genotype determination) is performed by comparing allele size (relative to an internal size standard) to a commercially AMEL provided STRD 5 S 818 kit allelic ladder FGA with calibrated repeat numbers (sized PET (red) according to the same internal size standard) GS 500 LIZ size standard (not shown above) LIZ (orange) An internal size standard is run with each sample and external standard to correlate sizes.

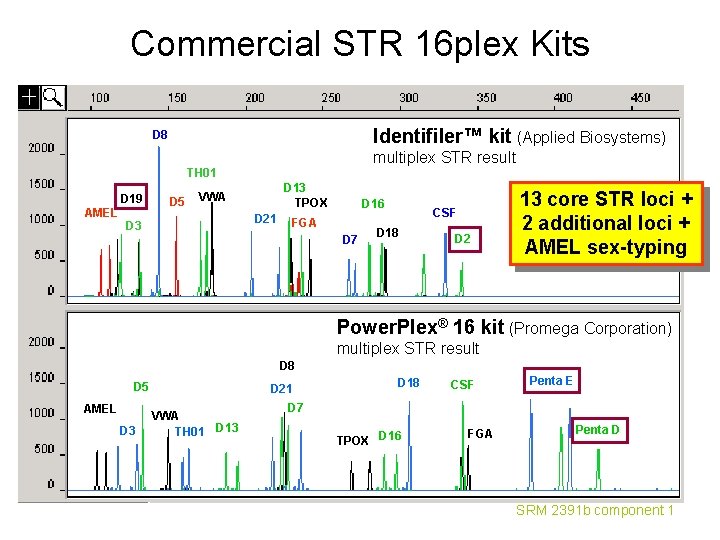
Commercial STR 16 plex Kits Identifiler™ kit (Applied Biosystems) D 8 multiplex STR result TH 01 D 19 AMEL D 5 D 13 TPOX VWA D 21 D 3 D 16 FGA D 7 CSF D 18 D 2 13 core STR loci + 2 additional loci + AMEL sex-typing Power. Plex® 16 kit (Promega Corporation) multiplex STR result D 8 D 5 AMEL D 3 D 21 VWA TH 01 D 13 D 18 CSF Penta E D 7 TPOX D 16 FGA Penta D SRM 2391 b component 1

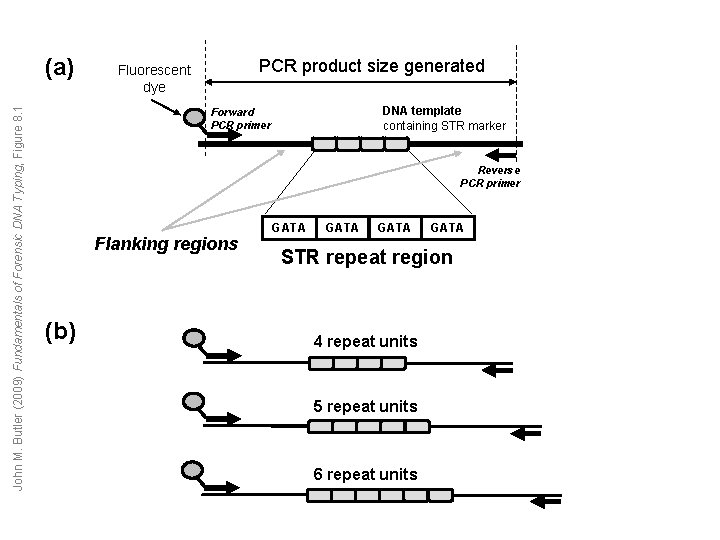
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 1 (a) PCR product size generated Fluorescent dye DNA template containing STR marker Forward PCR primer Reverse PCR primer Flanking regions (b) GATA STR repeat region 4 repeat units 5 repeat units 6 repeat units

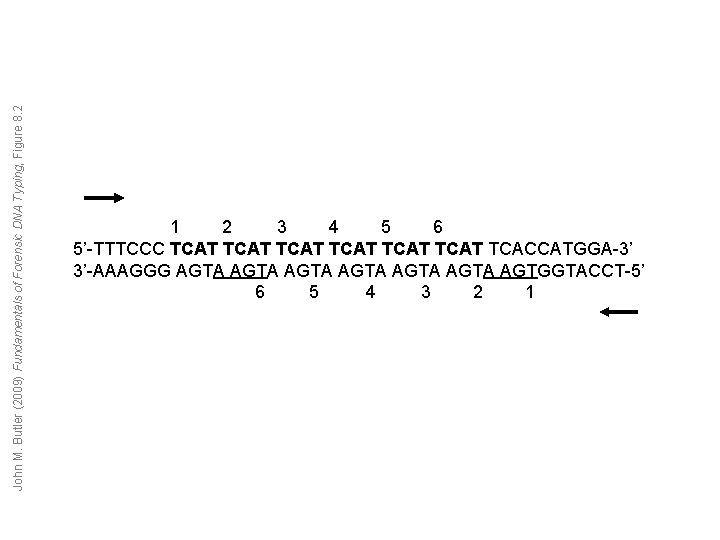
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 2 1 2 3 4 5 6 5’-TTTCCC TCAT TCAT TCACCATGGA-3’ 3’-AAAGGG AGTA AGTA AGTGGTACCT-5’ 6 5 4 3 2 1

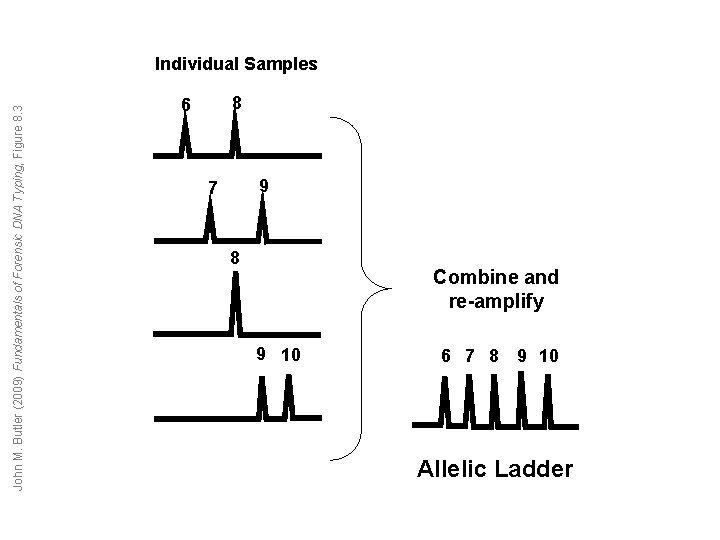
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 3 Individual Samples 8 6 9 7 8 Combine and re-amplify 9 10 6 7 8 9 10 Allelic Ladder

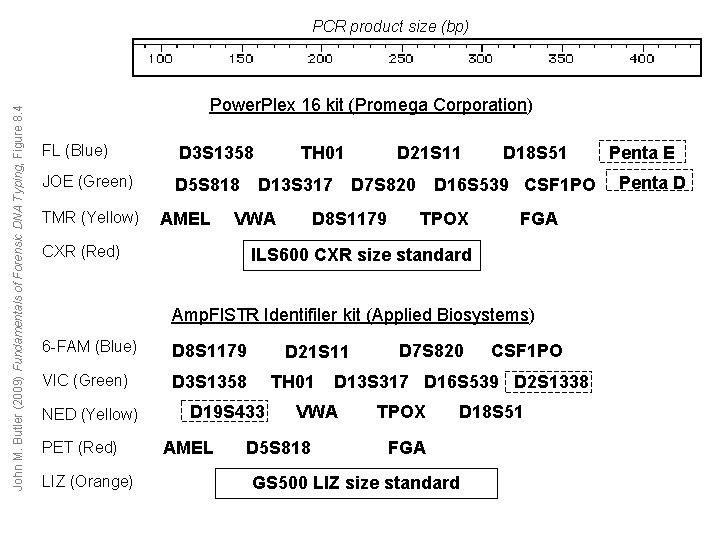
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 4 PCR product size (bp) Power. Plex 16 kit (Promega Corporation) FL (Blue) D 3 S 1358 JOE (Green) D 5 S 818 TMR (Yellow) AMEL TH 01 D 13 S 317 VWA CXR (Red) D 21 S 11 D 7 S 820 D 8 S 1179 D 18 S 51 D 16 S 539 CSF 1 PO TPOX FGA ILS 600 CXR size standard Amp. Fl. STR Identifiler kit (Applied Biosystems) 6 -FAM (Blue) D 8 S 1179 VIC (Green) D 3 S 1358 NED (Yellow) PET (Red) LIZ (Orange) D 21 S 11 TH 01 D 19 S 433 AMEL CSF 1 PO D 13 S 317 D 16 S 539 D 2 S 1338 VWA D 5 S 818 D 7 S 820 TPOX D 18 S 51 FGA GS 500 LIZ size standard Penta E Penta D

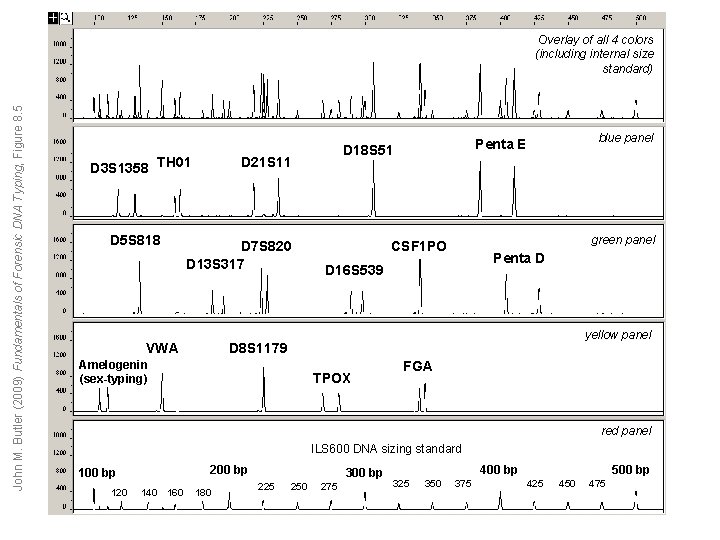
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 5 Overlay of all 4 colors (including internal size standard) D 3 S 1358 TH 01 D 5 S 818 D 21 S 11 D 7 S 820 D 13 S 317 VWA blue panel Penta E D 18 S 51 green panel CSF 1 PO Penta D D 16 S 539 yellow panel D 8 S 1179 Amelogenin (sex-typing) TPOX FGA red panel ILS 600 DNA sizing standard 200 bp 120 140 160 180 300 bp 225 250 275 400 bp 325 350 375 500 bp 425 450 475

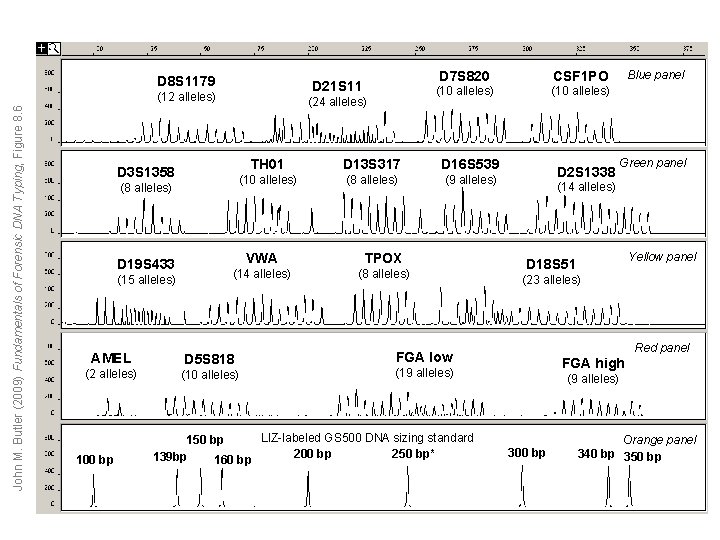
D 8 S 1179 D 21 S 11 John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 6 (12 alleles) (24 alleles) D 3 S 1358 (8 alleles) D 19 S 433 (15 alleles) CSF 1 PO (10 alleles) TH 01 D 13 S 317 D 16 S 539 (10 alleles) (8 alleles) (9 alleles) VWA TPOX (14 alleles) (8 alleles) AMEL D 5 S 818 (2 alleles) (10 alleles) 100 bp D 7 S 820 D 2 S 1338 Green panel (14 alleles) Yellow panel D 18 S 51 (23 alleles) Red panel FGA low FGA high (19 alleles) LIZ-labeled GS 500 DNA sizing standard 150 bp 200 bp 250 bp* 139 bp 160 bp Blue panel (9 alleles) 300 bp Orange panel 340 bp 350 bp

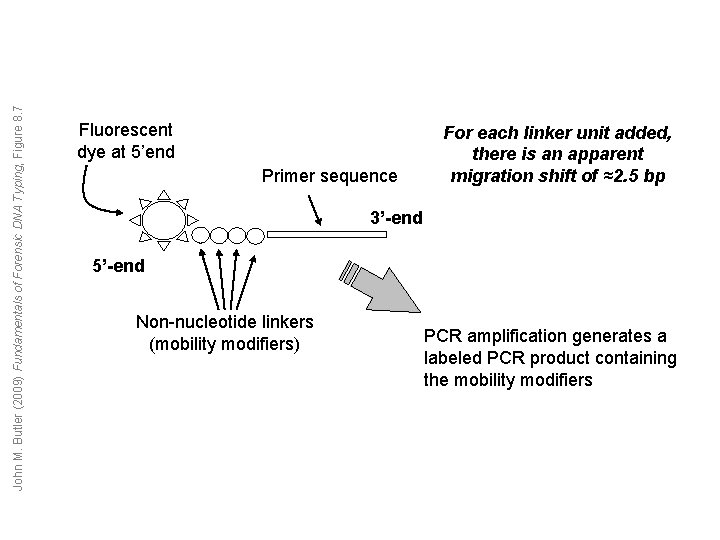
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 7 Fluorescent dye at 5’end Primer sequence For each linker unit added, there is an apparent migration shift of ≈2. 5 bp 3’-end 5’-end Non-nucleotide linkers (mobility modifiers) PCR amplification generates a labeled PCR product containing the mobility modifiers

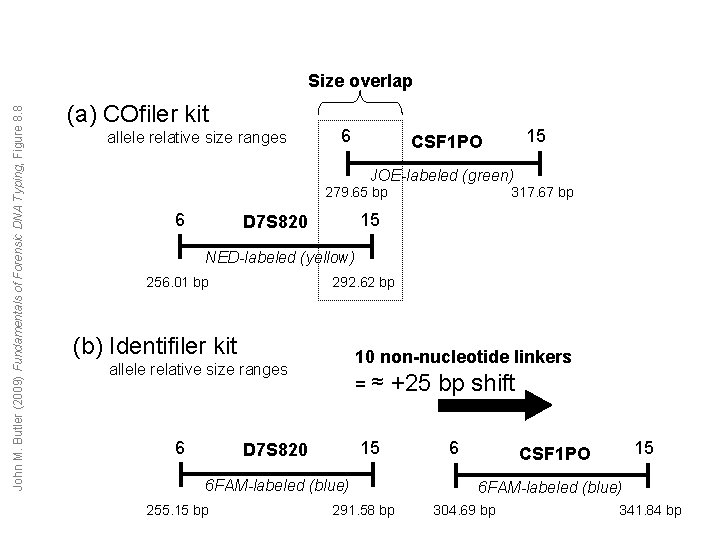
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 8 Size overlap (a) COfiler kit allele relative size ranges 6 15 CSF 1 PO JOE-labeled (green) 279. 65 bp 6 317. 67 bp 15 D 7 S 820 NED-labeled (yellow) 256. 01 bp 292. 62 bp (b) Identifiler kit 10 non-nucleotide linkers allele relative size ranges 6 D 7 S 820 =≈ +25 bp shift 15 6 6 FAM-labeled (blue) 255. 15 bp 291. 58 bp 15 CSF 1 PO 6 FAM-labeled (blue) 304. 69 bp 341. 84 bp

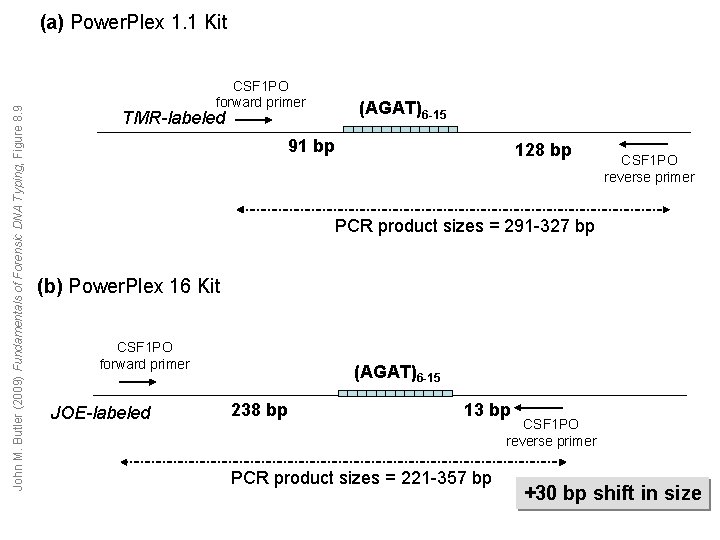
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 9 (a) Power. Plex 1. 1 Kit CSF 1 PO forward primer (AGAT)6 -15 TMR-labeled 91 bp 128 bp CSF 1 PO reverse primer PCR product sizes = 291 -327 bp (b) Power. Plex 16 Kit CSF 1 PO forward primer JOE-labeled (AGAT)6 -15 238 bp 13 bp PCR product sizes = 221 -357 bp CSF 1 PO reverse primer +30 bp shift in size

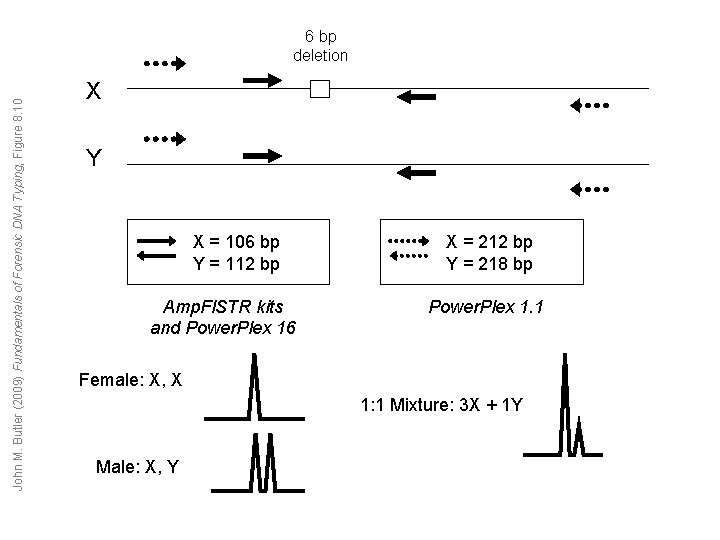
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 10 6 bp deletion X Y X = 106 bp Y = 112 bp Amp. Fl. STR kits and Power. Plex 16 X = 212 bp Y = 218 bp Power. Plex 1. 1 Female: X, X 1: 1 Mixture: 3 X + 1 Y Male: X, Y

John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 8. 11

TH 01 D 19 D 3 D 8 AMEL D 5 TPOX VWA D 7 D 13 D 21 CSF D 16 D 2 D 18 FGA DNA Size (bp) D 8 S 1179 6 FAM (blue) D 7 S 820 D 21 S 11 CSF 1 PO TH 01 VIC (green) D 3 S 1358 D 13 S 317 D 2 S 1338 D 16 S 539 TPOX NED (yellow) D 19 S 433 AMEL PET (red) VWA D 5 S 818 D 18 S 51 FGA GS 500 LIZ size standard LIZ (orange) 1 in 837 trillion (probability of this profile occurring at random)

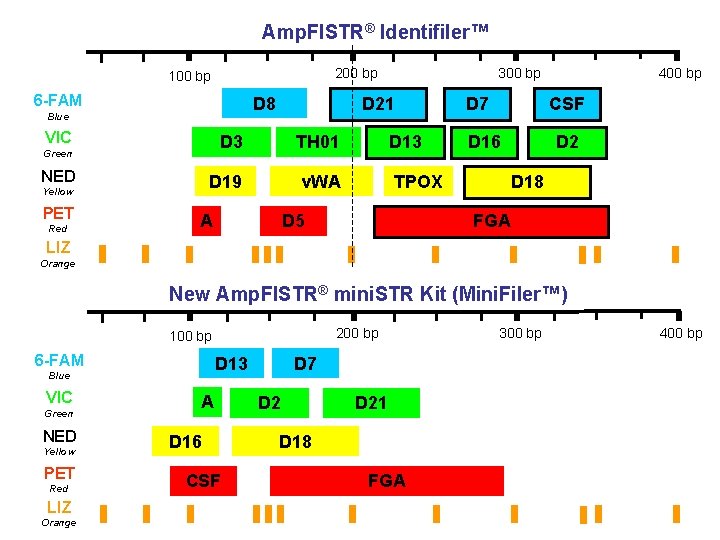
Amp. Fl. STR® Identifiler™ 200 bp 100 bp 6 -FAM D 8 Blue VIC Yellow PET Red D 21 D 3 TH 01 D 13 D 19 v. WA TPOX Green NED 300 bp A D 5 D 7 400 bp CSF D 16 D 2 D 18 FGA LIZ Orange New Amp. Fl. STR® mini. STR Kit (Mini. Filer™) 200 bp 100 bp 6 -FAM D 13 Blue VIC Green NED Yellow PET Red LIZ Orange A D 16 CSF D 7 D 21 D 18 FGA 300 bp 400 bp

Chapter 8 – Points for Discussion • Why are tetranucleotide repeat loci preferred over dinucleotide repeat loci in forensic testing? • What two innovations are used in the Identifiler kit to enable 16 loci to be simultaneously amplified and detected in a single reaction? • Why is it valuable to include the sex-typing marker amelogenin in STR typing kits?