Chapter 7 RNA and Protein Structure Structure Prediction


- Slides: 21

Chapter 7 RNA and Protein Structure: Structure Prediction

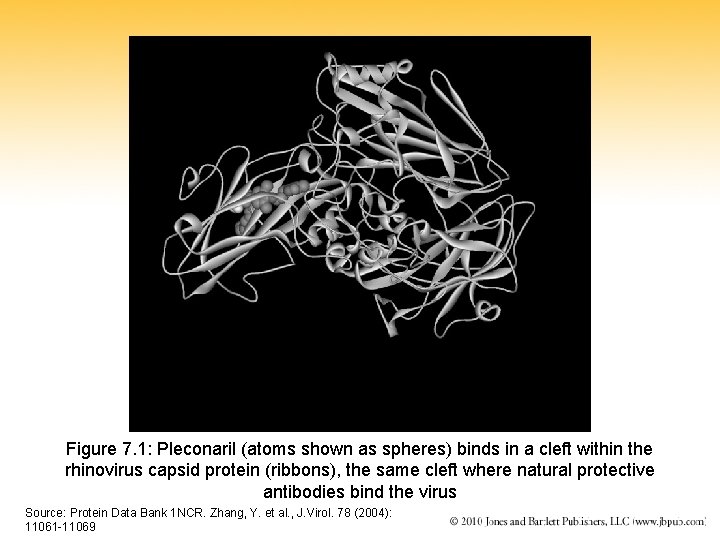
Figure 7. 1: Pleconaril (atoms shown as spheres) binds in a cleft within the rhinovirus capsid protein (ribbons), the same cleft where natural protective antibodies bind the virus Source: Protein Data Bank 1 NCR. Zhang, Y. et al. , J. Virol. 78 (2004): 11061 -11069

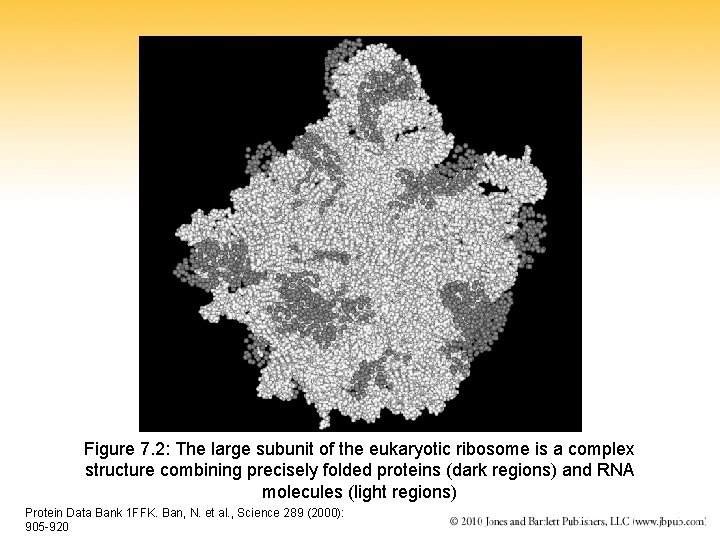
Figure 7. 2: The large subunit of the eukaryotic ribosome is a complex structure combining precisely folded proteins (dark regions) and RNA molecules (light regions) Protein Data Bank 1 FFK. Ban, N. et al. , Science 289 (2000): 905 -920

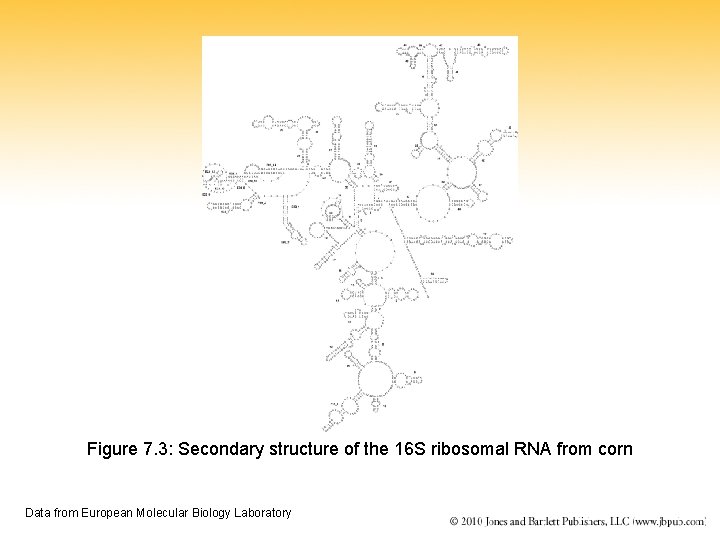
Figure 7. 3: Secondary structure of the 16 S ribosomal RNA from corn Data from European Molecular Biology Laboratory

Figure 7. 4: An alpha-helix as seen from the top Protein Data Bank 2 ZNA. Wang, A. H. -J. , Science 211 (1981): 171 -176

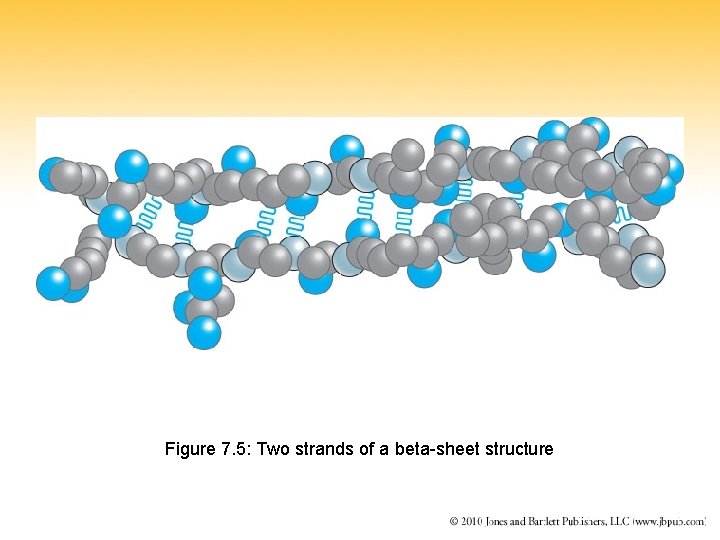
Figure 7. 5: Two strands of a beta-sheet structure

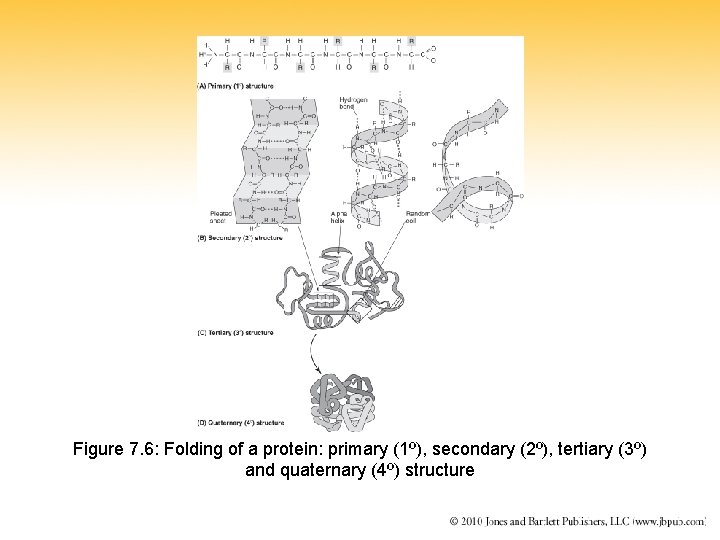
Figure 7. 6: Folding of a protein: primary (1º), secondary (2º), tertiary (3º) and quaternary (4º) structure

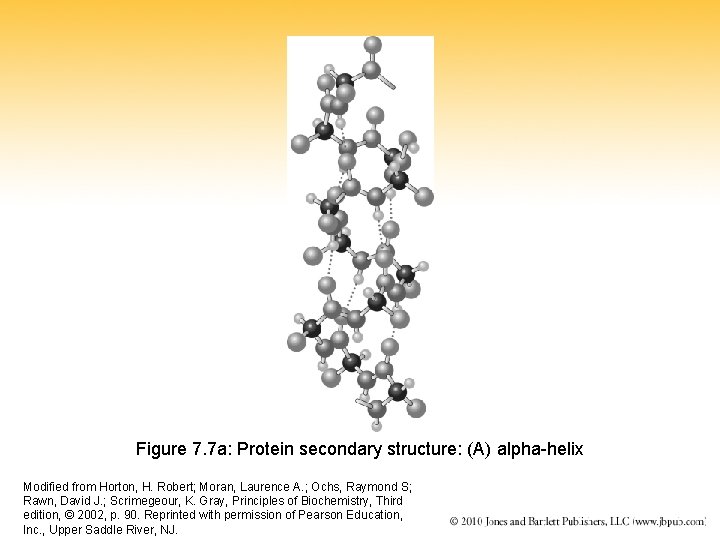
Figure 7. 7 a: Protein secondary structure: (A) alpha-helix Modified from Horton, H. Robert; Moran, Laurence A. ; Ochs, Raymond S; Rawn, David J. ; Scrimegeour, K. Gray, Principles of Biochemistry, Third edition, © 2002, p. 90. Reprinted with permission of Pearson Education, Inc. , Upper Saddle River, NJ.

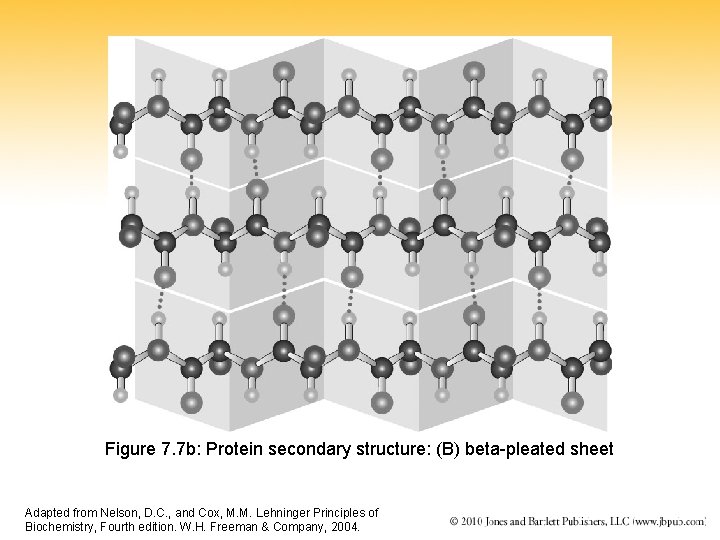
Figure 7. 7 b: Protein secondary structure: (B) beta-pleated sheet Adapted from Nelson, D. C. , and Cox, M. M. Lehninger Principles of Biochemistry, Fourth edition. W. H. Freeman & Company, 2004.

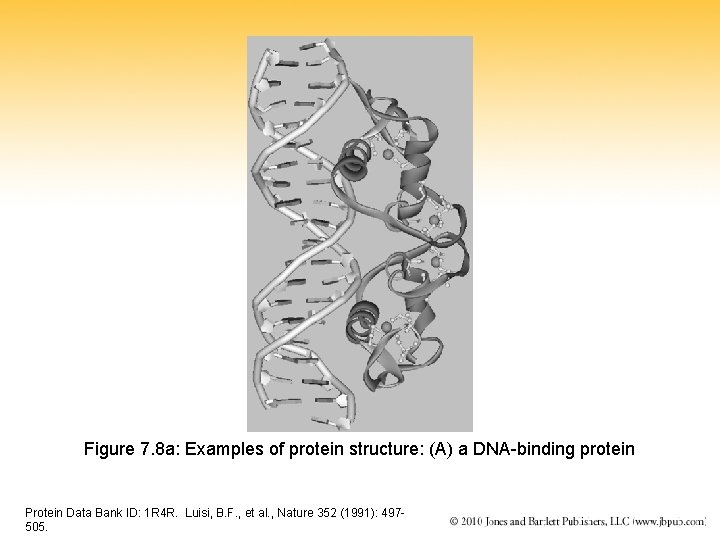
Figure 7. 8 a: Examples of protein structure: (A) a DNA-binding protein Protein Data Bank ID: 1 R 4 R. Luisi, B. F. , et al. , Nature 352 (1991): 497505.

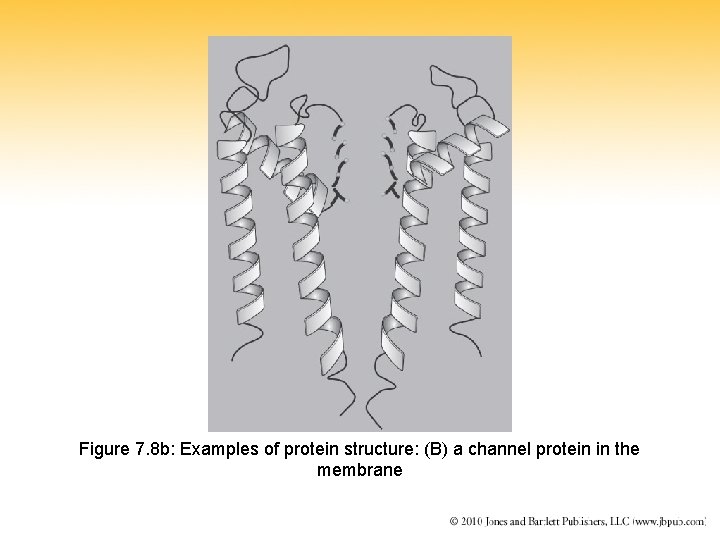
Figure 7. 8 b: Examples of protein structure: (B) a channel protein in the membrane

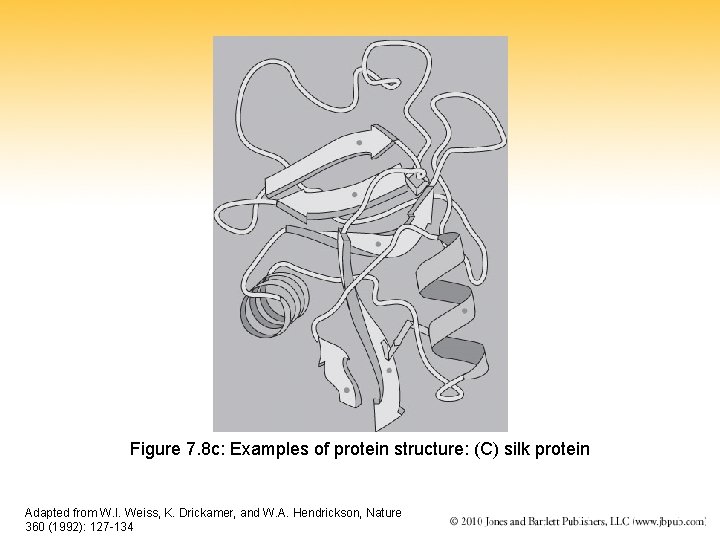
Figure 7. 8 c: Examples of protein structure: (C) silk protein Adapted from W. I. Weiss, K. Drickamer, and W. A. Hendrickson, Nature 360 (1992): 127 -134

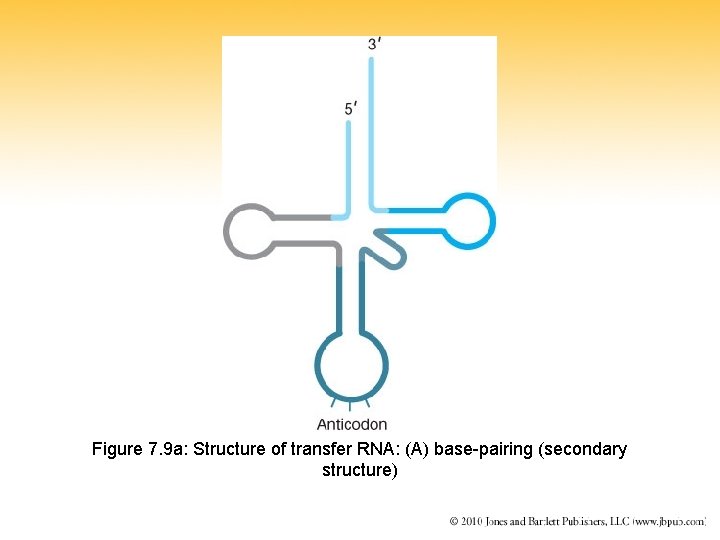
Figure 7. 9 a: Structure of transfer RNA: (A) base-pairing (secondary structure)

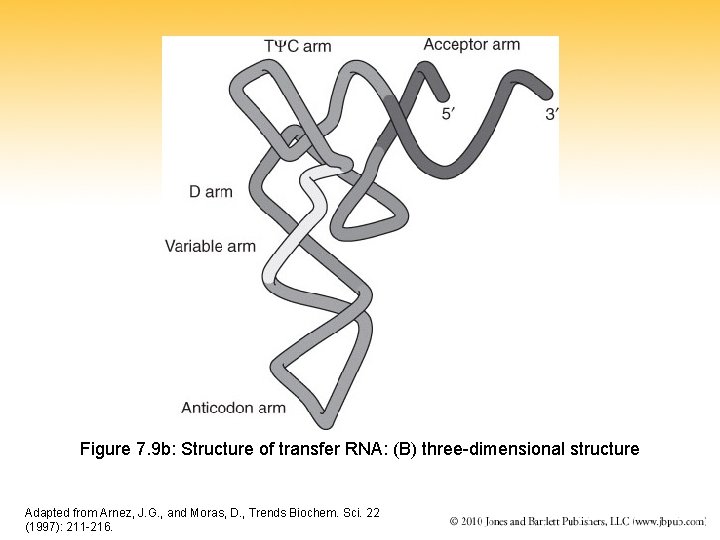
Figure 7. 9 b: Structure of transfer RNA: (B) three-dimensional structure Adapted from Arnez, J. G. , and Moras, D. , Trends Biochem. Sci. 22 (1997): 211 -216.

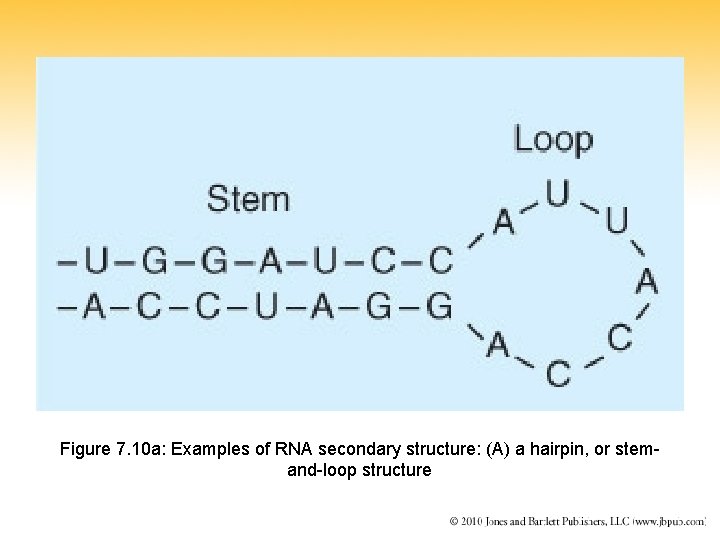
Figure 7. 10 a: Examples of RNA secondary structure: (A) a hairpin, or stemand-loop structure

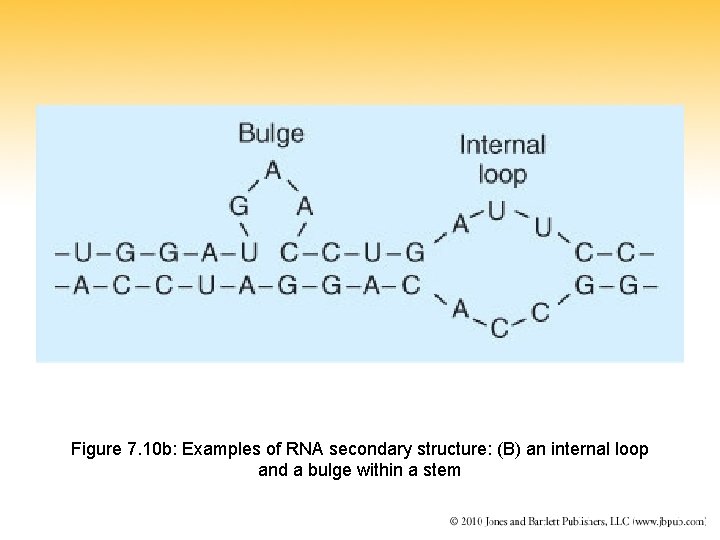
Figure 7. 10 b: Examples of RNA secondary structure: (B) an internal loop and a bulge within a stem

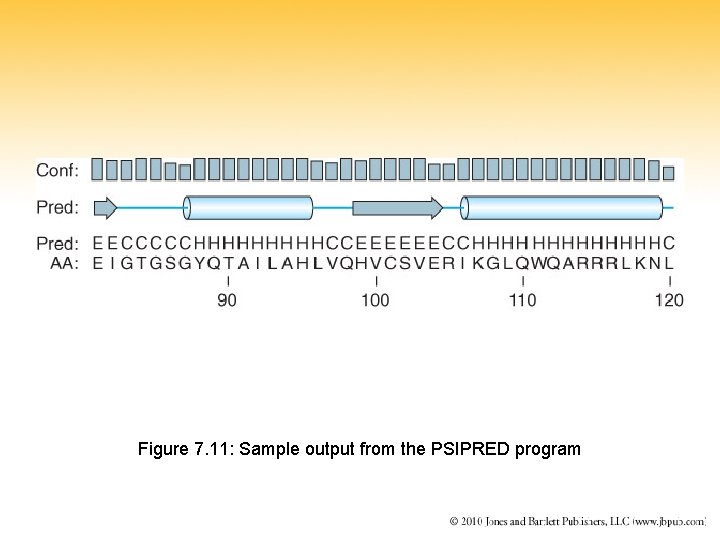
Figure 7. 11: Sample output from the PSIPRED program

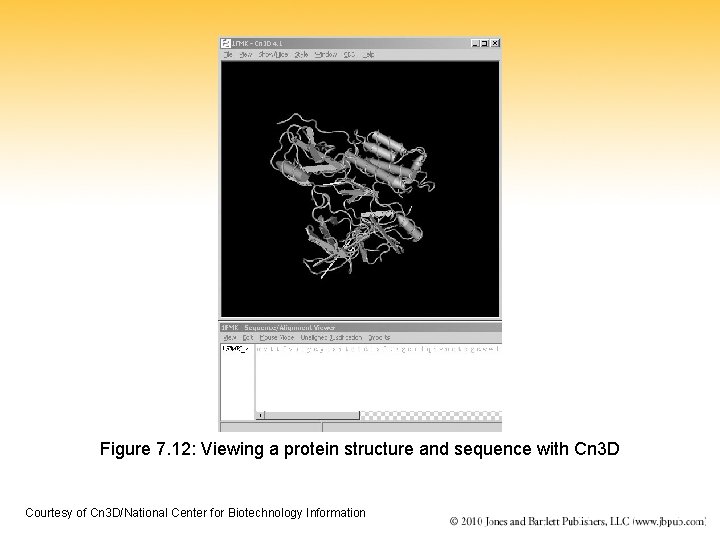
Figure 7. 12: Viewing a protein structure and sequence with Cn 3 D Courtesy of Cn 3 D/National Center for Biotechnology Information

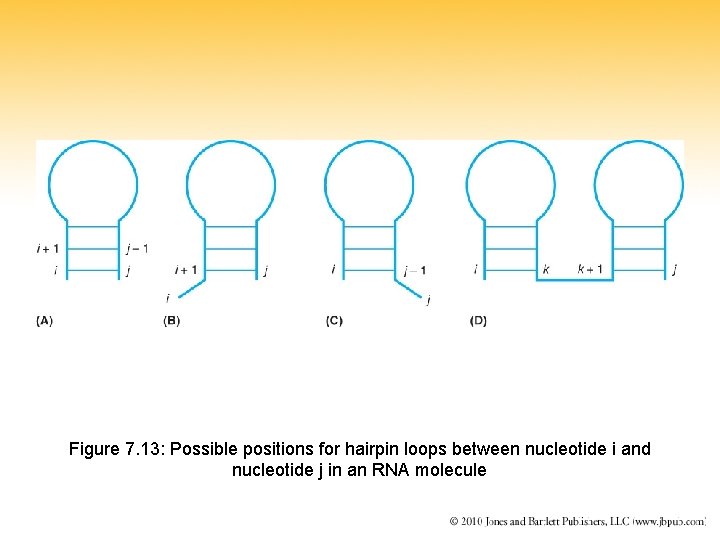
Figure 7. 13: Possible positions for hairpin loops between nucleotide i and nucleotide j in an RNA molecule

Figure 7. 14: Screenshot from a computer participating in the Folding@home project Courtesy of Vijay Pande, Stanford University and Folding@home

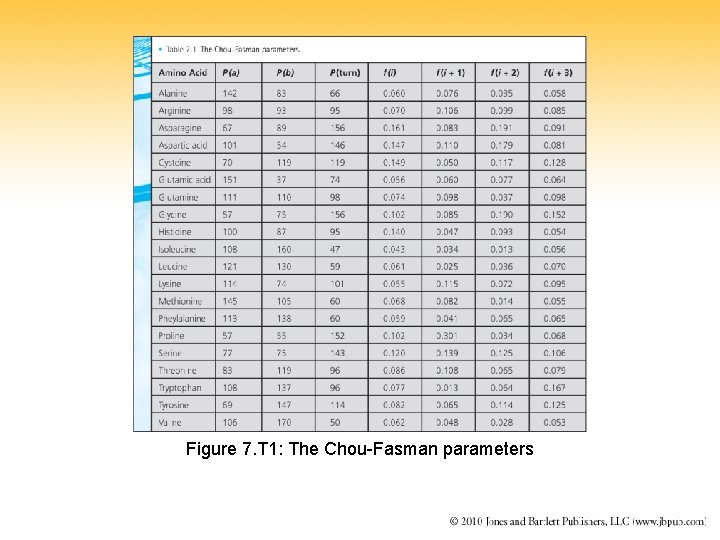
Figure 7. T 1: The Chou-Fasman parameters