Chapter 7 I Introduction to Enzyme 1 Overview




![1] Enzymes are usually very large in comparison with substrate : n So only 1] Enzymes are usually very large in comparison with substrate : n So only](https://slidetodoc.com/presentation_image_h/e37d59c9148d3f22eb341cd3f17163a4/image-20.jpg)
![2] Active site is a 3 -dimensional entity (3 -D) q q Not a 2] Active site is a 3 -dimensional entity (3 -D) q q Not a](https://slidetodoc.com/presentation_image_h/e37d59c9148d3f22eb341cd3f17163a4/image-22.jpg)
![3] Substrate is bound by relatively weak forces n When there is steric complementary 3] Substrate is bound by relatively weak forces n When there is steric complementary](https://slidetodoc.com/presentation_image_h/e37d59c9148d3f22eb341cd3f17163a4/image-24.jpg)


- Slides: 35

Chapter 7 I. Introduction to Enzyme

1) Overview Ø All reactions in body are mediated by enzymes, which are protein catalysts that increase that rate of reactions without being changed in overall process. Ø Enzymes accelerate reactions by factors of at least a million Ø All known enzymes are proteins Ø The most striking characteristics of enzymes are their catalytic power and specificity Ø The action of many enzymes are regulated.

2) Nomeculture Ø Each enzyme is assigned two names Ø The first is its short, recommended name, convenient for everyday use. Ø The second is the more complete systematic name, which is used when enzyme must be identified without ambiguity.

A. Recommended name Ø Most commonly used enzyme names have the suffix “ -ase” attached: Ø To the substrate of the reaction (for example, glucosidase, urease, sucrase) Ø To a description of the action performed (for example, lactate dehydorgenase and adenylyl cyclase) Ø Some enzymes retain their original trivial names, which give no hint of the associated enzymic reaction, of example, trypsin and pepsin.

B. Systematic name Ø The International Union of Biochemistry and Molecular Biology (IUBM) developed a system of nomenclature in which enzymes are divides into six major classes: 1. Oxidoreductase: Catalyze oxidation-reduction reactions. 2. Transferases : Catalyze transfer of C-, N-, or Pcontaining groups 3. Hydrolases : Catalyze cleavage of bonds by addition of water

4. Lyases : Catalyze cleavage of C-C, C-S and certain C-N bonds 5. Isomerases : Catalyze racemization of optical or geometric isomers 6. Ligases : Catalyze formation of bonds between carbon and O, S, N coupled to hydrolysis of high energy phosphates Ø The suffix –ase is attached to a fairly complete description of the chemical reaction catalyzed.

3) Properties of Enzymes v Enzymes are • Protein catalysts that increase the velocity of a chemical reaction • NOT consumed during the reaction they catalyze. v Some types of RNA can act like enzymes, usually catalyzing the cleavage and synthesis of phosphodiester bonds. v RNA’s with catalytic activity are called ribozymes

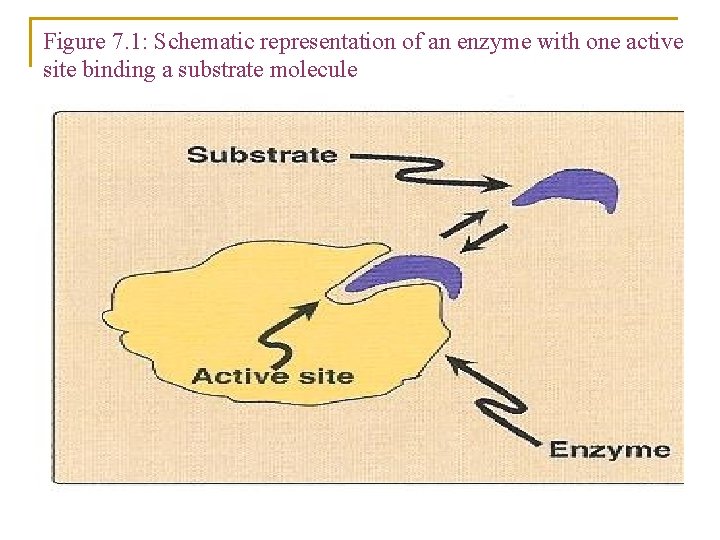
A. Active sites n n n Enzyme molecules contain a special pocket or cleft called the active site. The active site contains amino acid side chains that create a three-dimensional surface complementary to the substrate (Figure 7. 1). The active site binds the substrate, forming an enzyme-substrate (ES) complex. ES is converted to enzyme-product (EP), which subsequently dissociates to enzymes and product.

Figure 7. 1: Schematic representation of an enzyme with one active site binding a substrate molecule

B. Catalytic efficiency n Most enzyme- catalyzed reactions are highly efficient, proceeding from 103 to 108 times faster then uncataluzed reactions. n Each enzyme molecule is capable of transforming 100 to 1000 sub-molecules of substrate converted to product per enzyme molecule per second is called the turnover number

C. Specificity n Enzymes are highly specific: Ø Interacting with one or few substrates Ø Catalyzing only one type of chemical reaction.

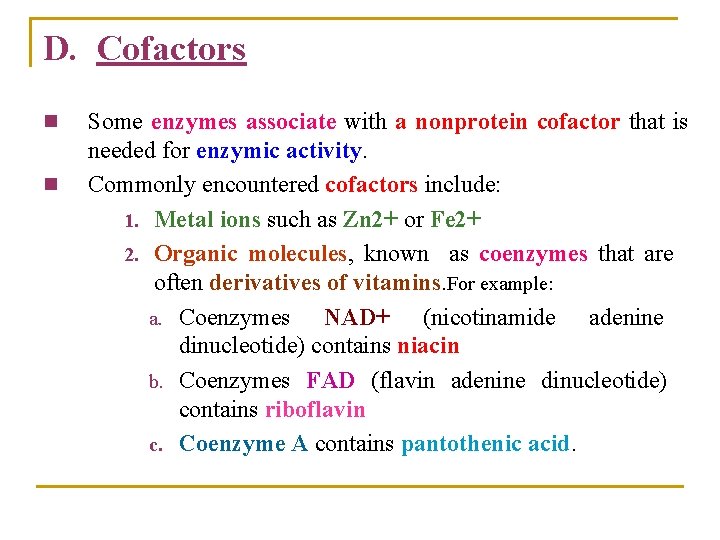
D. Cofactors n n Some enzymes associate with a nonprotein cofactor that is needed for enzymic activity. Commonly encountered cofactors include: 1. Metal ions such as Zn 2+ or Fe 2+ 2. Organic molecules, known as coenzymes that are often derivatives of vitamins. For example: a. Coenzymes NAD+ (nicotinamide adenine dinucleotide) contains niacin b. Coenzymes FAD (flavin adenine dinucleotide) contains riboflavin c. Coenzyme A contains pantothenic acid.

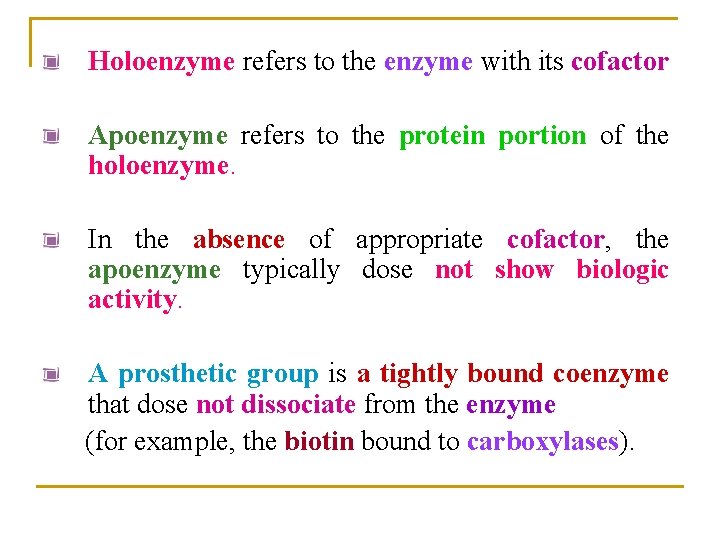
Holoenzyme refers to the enzyme with its cofactor Apoenzyme refers to the protein portion of the holoenzyme. In the absence of appropriate cofactor, the apoenzyme typically dose not show biologic activity. A prosthetic group is a tightly bound coenzyme that dose not dissociate from the enzyme (for example, the biotin bound to carboxylases).

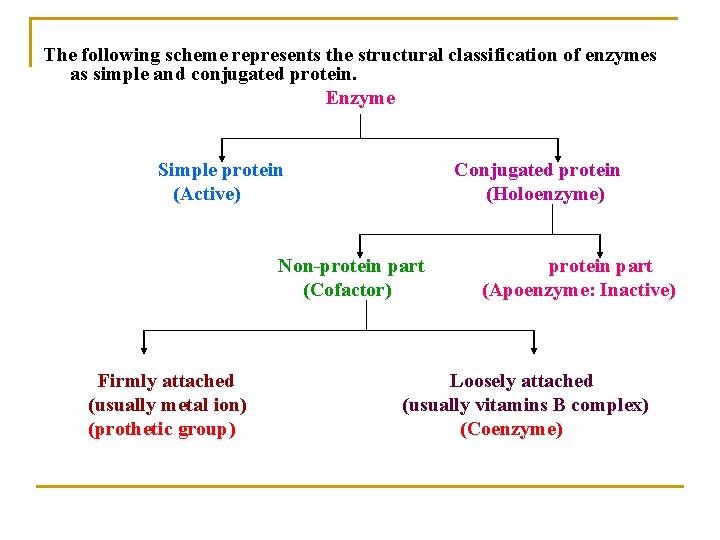
The following scheme represents the structural classification of enzymes as simple and conjugated protein. Enzyme Simple protein (Active) Conjugated protein (Holoenzyme) Non-protein part (Cofactor) Firmly attached (usually metal ion) (prothetic group) protein part (Apoenzyme: Inactive) Loosely attached (usually vitamins B complex) (Coenzyme)

E. Regulation n Enzyme activity can be regulated, that is, enzymes can be: § Activated § Inhibited § So that the rate of Product formation responds to the needs of the cell.

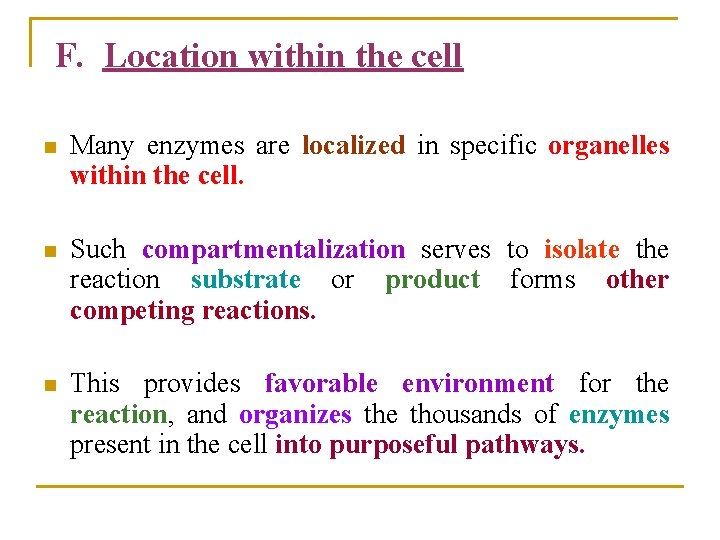
F. Location within the cell n Many enzymes are localized in specific organelles within the cell. n Such compartmentalization serves to isolate the reaction substrate or product forms other competing reactions. n This provides favorable environment for the reaction, and organizes the thousands of enzymes present in the cell into purposeful pathways.

Figure 7. 2: The intracellular location of some important biochemical pathways

4) How Enzyme Work n The mechanism of enzyme action can be viewed form the perspective that describes how the active site chemically facilitates catalysis.

Active site n n n The active site of an enzyme is the region that binds the substrate So the active site contributes the amino acid residues that directly participate in the making and breaking of chemical bonds. The amino acid residues are called the catalytic groups (of enzyme). Enzymes differ widely in n Structure n Function n Mode of catalysis So active sites vary, but possible to make some generalizations:
![1 Enzymes are usually very large in comparison with substrate n So only 1] Enzymes are usually very large in comparison with substrate : n So only](https://slidetodoc.com/presentation_image_h/e37d59c9148d3f22eb341cd3f17163a4/image-20.jpg)
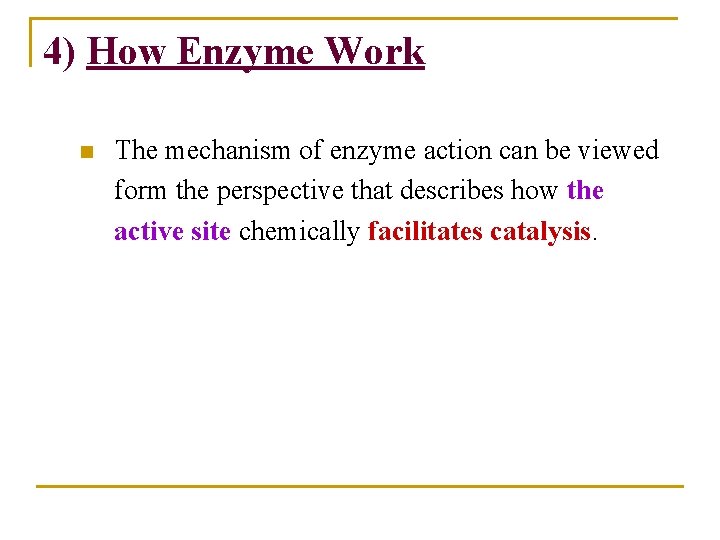
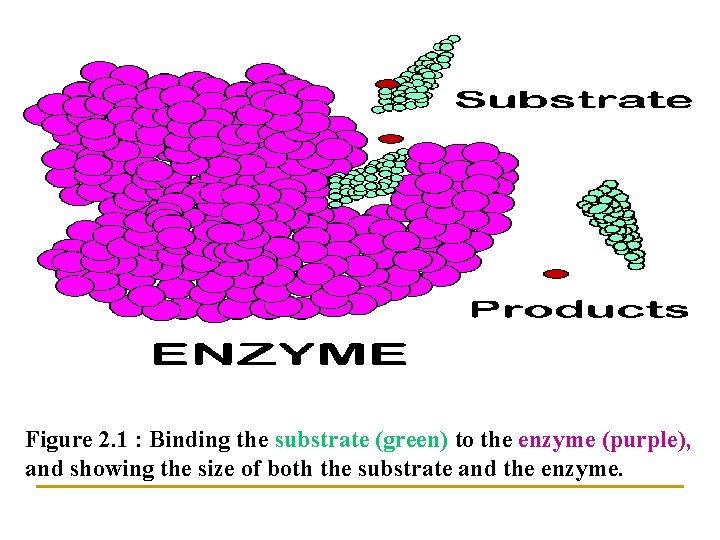
1] Enzymes are usually very large in comparison with substrate : n So only small portion of amino acid residues are n Near n In direct contact with substrate n Evidence for this includes the observation that: n Large portions of certain enzymes may be removed without loss of catalytic activity (see Figure 2. 1).

Figure 2. 1 : Binding the substrate (green) to the enzyme (purple), and showing the size of both the substrate and the enzyme.
![2 Active site is a 3 dimensional entity 3 D q q Not a 2] Active site is a 3 -dimensional entity (3 -D) q q Not a](https://slidetodoc.com/presentation_image_h/e37d59c9148d3f22eb341cd3f17163a4/image-22.jpg)
2] Active site is a 3 -dimensional entity (3 -D) q q Not a point or a plane, usually an intricate (complex) pocket or cleft structurally designed to accept the structure of the substrate in 3 -D terms. The residues which constitute the active site are n Not close to each other in the primary sequence, n But the tertiary fold brings them close together in three-dimensional space.

n Active sites often involve residues on connecting loops between n -Helices n ß-Sheets n In many instances, active sites occur at the junction between two domains making tertiary contacts.
![3 Substrate is bound by relatively weak forces n When there is steric complementary 3] Substrate is bound by relatively weak forces n When there is steric complementary](https://slidetodoc.com/presentation_image_h/e37d59c9148d3f22eb341cd3f17163a4/image-24.jpg)
3] Substrate is bound by relatively weak forces n When there is steric complementary between the substrate and the active site of the enzyme. n Electrostatic n Hydrogen bonds n Van der Waals interactions all are important. The free energy of interaction between E and S ranges 12 to 36 k. J/mole comparing this with the strength of a covalent bond which reach up to 450 k. J/mole.

5) Factors Affecting Reaction Velocity n n Enzyme can be isolated from cells, and their properties studied in a test tube (that is, in vitro). Different enzymes show different responses to changes in substrate concentration, temperature, and p. H. This section describes factors that influence the reaction velocity of enzymes. Enzymic responses to these factors give us valuable clues as to how enzymes function in living cells.

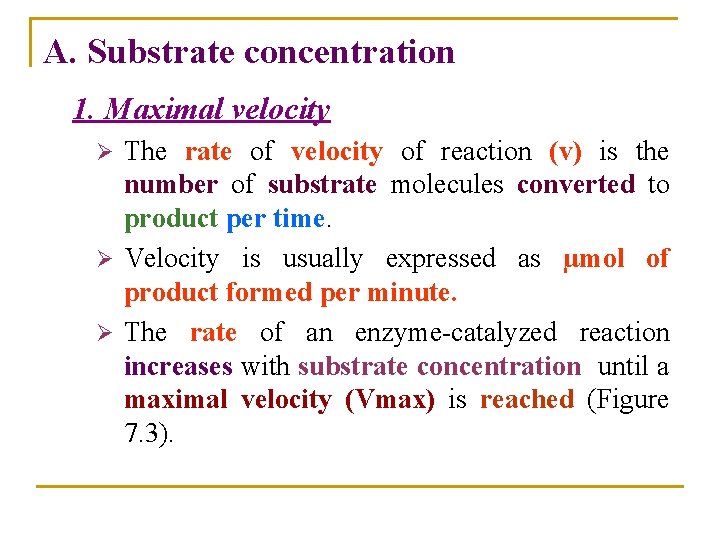
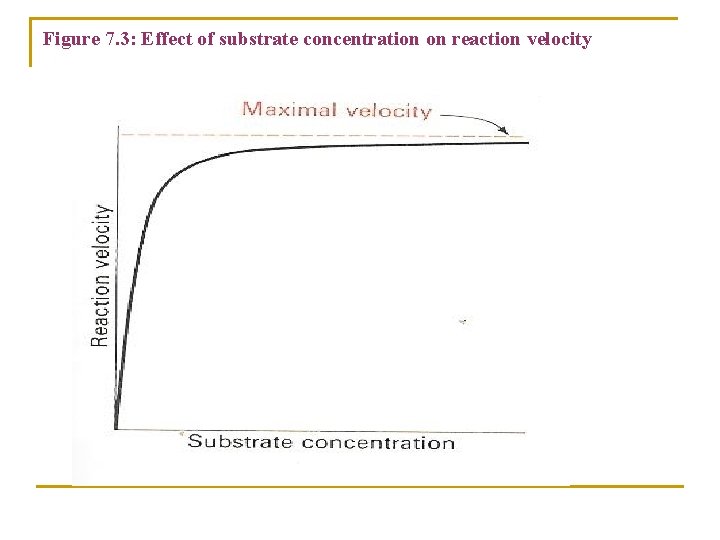
A. Substrate concentration 1. Maximal velocity The rate of velocity of reaction (v) is the number of substrate molecules converted to product per time. Ø Velocity is usually expressed as µmol of product formed per minute. Ø The rate of an enzyme-catalyzed reaction increases with substrate concentration until a maximal velocity (Vmax) is reached (Figure 7. 3). Ø

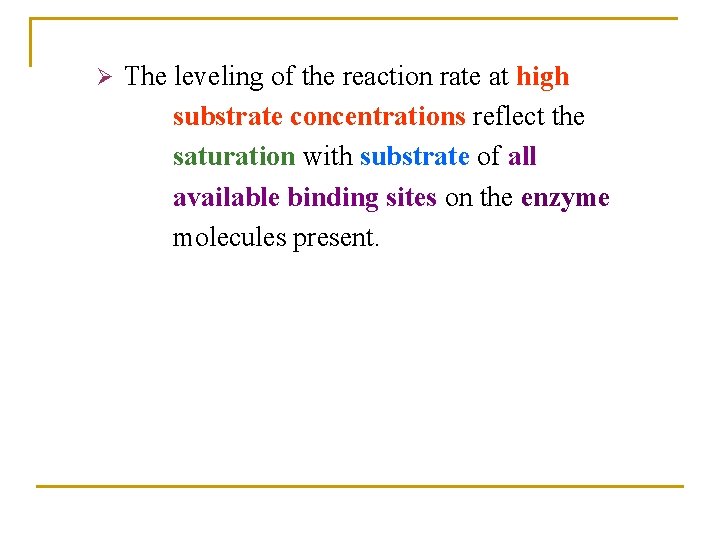
Ø The leveling of the reaction rate at high substrate concentrations reflect the saturation with substrate of all available binding sites on the enzyme molecules present.

Figure 7. 3: Effect of substrate concentration on reaction velocity

2. Hyperbolic shape of the enzyme kinetics curve Ø Most enzymes show Michaelis-Menten kinetics. Ø In which the plot of initial reaction velocity, (v 0), against substrate concentration [S], is hyperbolic.

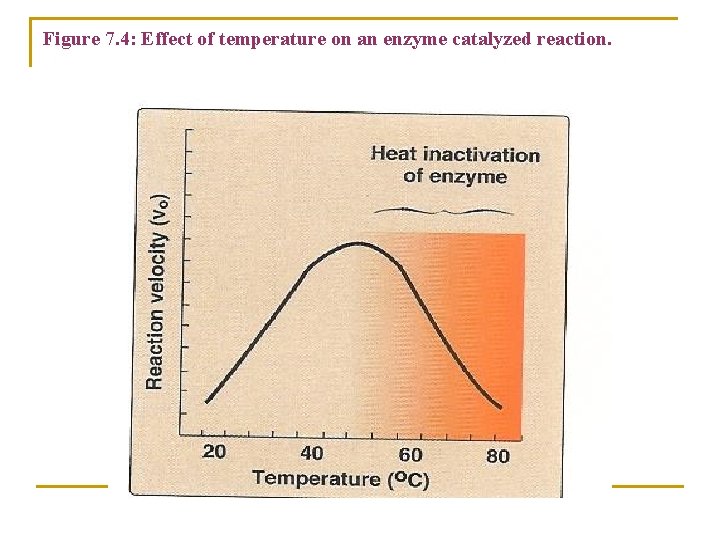
B. Temperature 1. Increase of velocity with temperature: ü The reaction velocity increases with temperature until a peak velocity is reached (Figure 7. 4). ü This increase in the velocity is the result of increased number of molecules that form the products of the reaction.

Figure 7. 4: Effect of temperature on an enzyme catalyzed reaction.

2. Decrease of velocity with higher temperature: ü Further elevation of the temperature results in a decrease in reaction velocity as a result of temperature-induced Denaturation of enzyme (see Figure 7. 4).

C. p. H 1. Effect of p. H on the ionization of the active site ü The concentration of H+ affects reaction velocity in several ways. ü First, the catalytic process usually requires that the enzyme and substrate have specific chemical groups in either an ionized or unionized state in order to interact. ü For example, catalytic activity may require that an amino group of the enzyme be in the protonated form (-NH 3+). At alkaline p. H, this group is deportonated, and the rate of reaction, therefore, declines.

2. Effect of p. H on enzyme Denaturation ü Extremes of p. H can also lead to denaturation of the enzyme ü Because the structure of the catalytically active protein molecule depends on the ionic character of the amino acid side chains.

3. The p. H optimum varies for different enzyme ü The p. H at which maximal enzyme activity is achieved is different for different enzymes, and often reflects the [H+] at which the enzyme functions in the body. ü For example, pepsin, a digestive enzyme in the stomach, is maximally active at p. H 2, whereas other enzymes, designed to work at neutral p. H are denatured by such an acidic environment.