Chapter 7 DNA Amplification Fundamentals of Forensic DNA



- Slides: 12

Chapter 7 DNA Amplification Fundamentals of Forensic DNA Typing Slides prepared by John M. Butler June 2009

Chapter 7 – DNA Amplification Chapter Summary Detection of minute amounts of biological material from crime scene evidence is possible due to DNA amplification via the polymerase chain reaction (PCR). Using multiplex PCR, specific regions of the human genome are simultaneously targeted with sequence-specific oligonucleotide PCR primers and copied with a DNA polymerase and deoxynucleotide triphosphate building blocks. Close to a billion copies of each specific region of the genome can be generated in a matter of a few hours by subjecting the DNA sample to typically 2832 cycles of heating and cooling that permit the replication process to occur. The PCR amplification process incorporates fluorescentlylabeled primers into the PCR products that enable multi-color fluorescence detection. In order to protect from contamination by post-PCR products, pre- and post-PCR steps are typically segregated in different laboratory space.

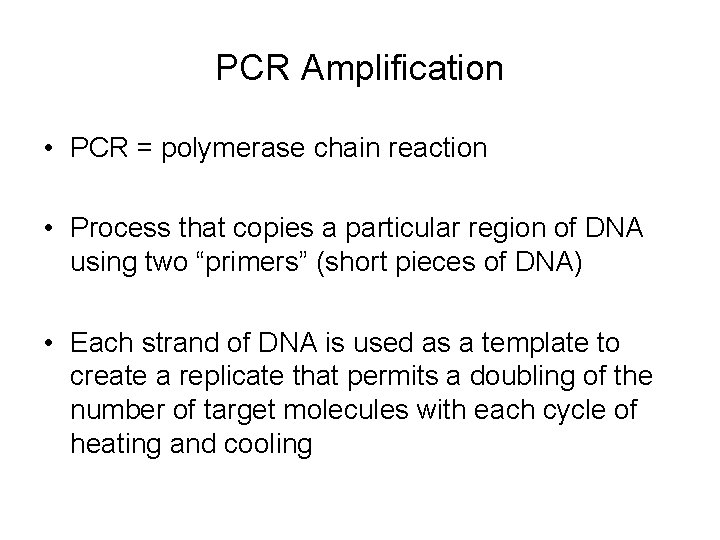
PCR Amplification • PCR = polymerase chain reaction • Process that copies a particular region of DNA using two “primers” (short pieces of DNA) • Each strand of DNA is used as a template to create a replicate that permits a doubling of the number of target molecules with each cycle of heating and cooling

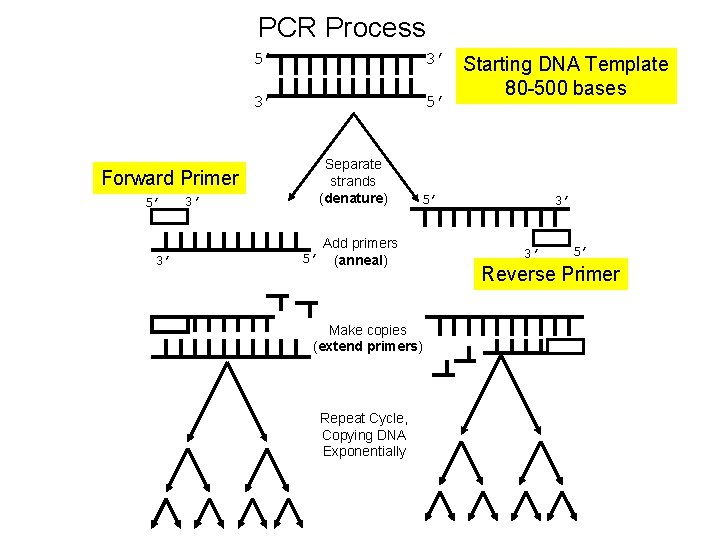
PCR Process 5’ 3’ Starting DNA Template 3’ 5’ Separate strands (denature) Forward Primer 5’ 3’ 3’ 5’ 5’ Add primers (anneal) Make copies (extend primers) Repeat Cycle, Copying DNA Exponentially 80 -500 bases 3’ 3’ 5’ Reverse Primer

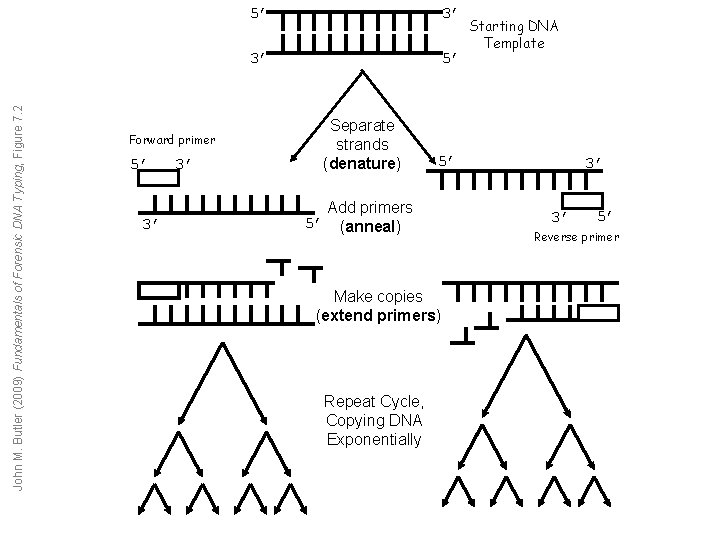
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 7. 2 Forward primer 5’ 3’ 3’ 5’ Separate strands (denature) 5’ Add primers 5’ (anneal) Make copies (extend primers) Repeat Cycle, Copying DNA Exponentially Starting DNA Template 3’ 3’ 5’ Reverse primer

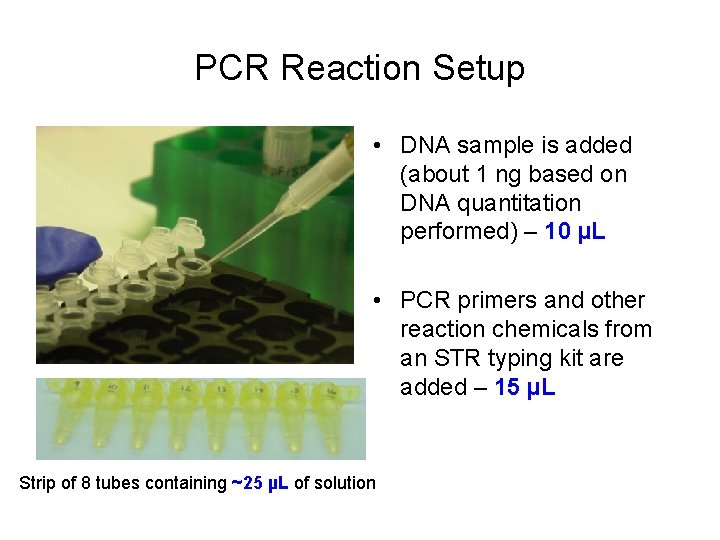
PCR Reaction Setup • DNA sample is added (about 1 ng based on DNA quantitation performed) – 10 µL • PCR primers and other reaction chemicals from an STR typing kit are added – 15 µL Strip of 8 tubes containing ~25 µL of solution

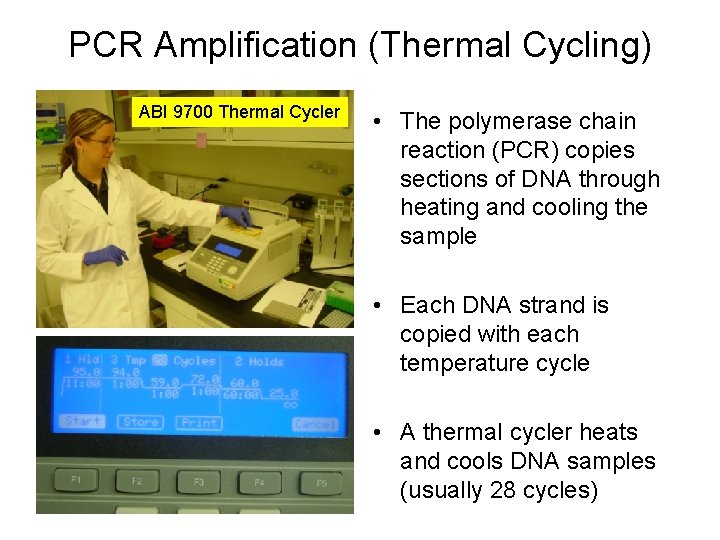
PCR Amplification (Thermal Cycling) ABI 9700 Thermal Cycler • The polymerase chain reaction (PCR) copies sections of DNA through heating and cooling the sample • Each DNA strand is copied with each temperature cycle • A thermal cycler heats and cools DNA samples (usually 28 cycles)

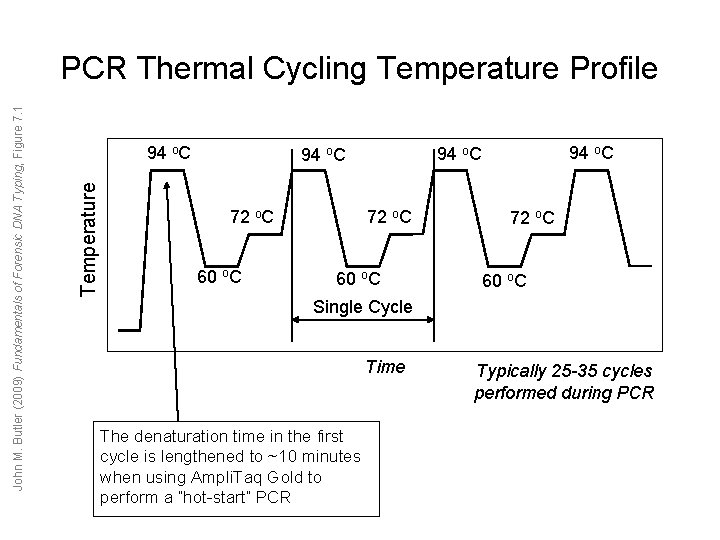
94 o. C Temperature John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 7. 1 PCR Thermal Cycling Temperature Profile 72 o. C 60 o. C 94 o. C 60 o. C 72 o. C 60 o. C Single Cycle Time The denaturation time in the first cycle is lengthened to ~10 minutes when using Ampli. Taq Gold to perform a “hot-start” PCR Typically 25 -35 cycles performed during PCR

John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 7. 3 Gene. Amp 9700 Thermal Cycler

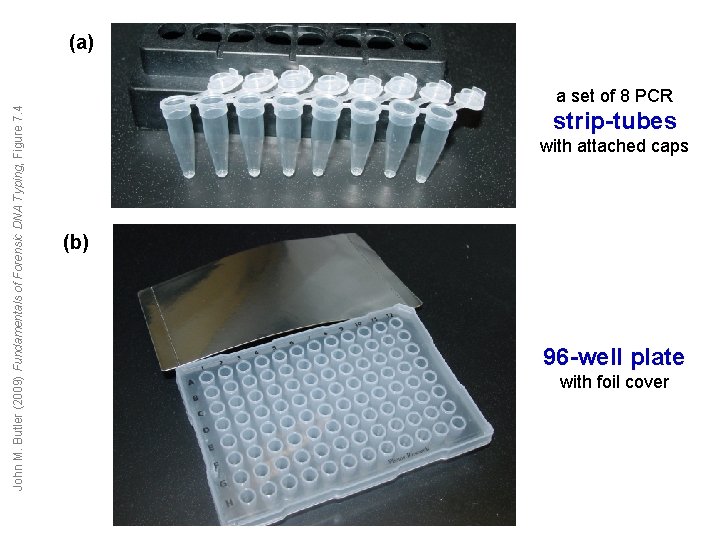
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 7. 4 (a) a set of 8 PCR strip-tubes with attached caps (b) 96 -well plate with foil cover

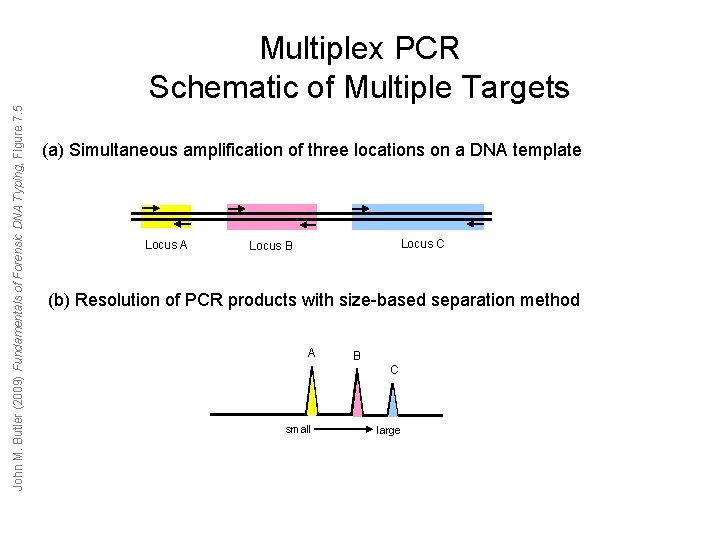
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 7. 5 Multiplex PCR Schematic of Multiple Targets (a) Simultaneous amplification of three locations on a DNA template Locus A Locus C Locus B (b) Resolution of PCR products with size-based separation method A B C small large

Chapter 7 – Points for Discussion • What challenges exist with designing multiplex PCR primers? • What are the advantages of a thermal stable, hot-start DNA polymerase? • Why is it important to separate pre-PCR and post-PCR processes? • What is the purpose of a negative amplification control? A postive amplification control? • What are some effective means to prevent contamination? • What are some effective means to clean up following a contamination event? • What are “stochastic effects” and why are they important to forensic DNA analysis?