Chapter 7 Analyzing DNA and gene structure variation




- Slides: 31

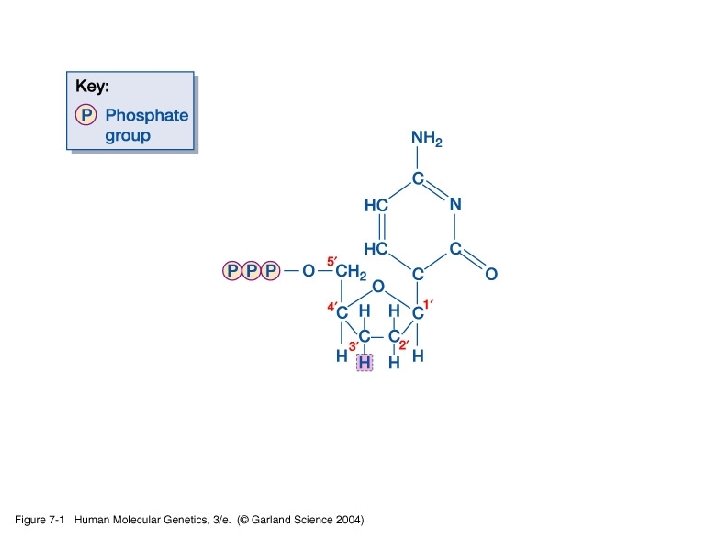
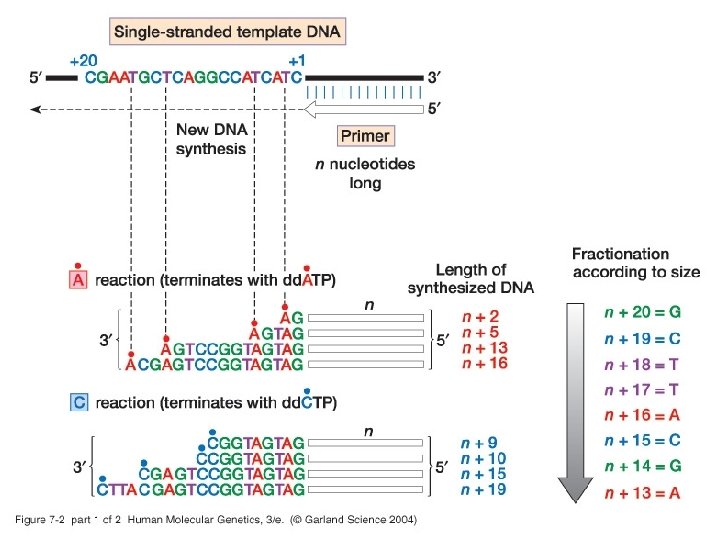
Chapter 7 Analyzing DNA and gene structure, variation and expression 1. Sequencing and genotyping DNA • Standard/manual DNA sequencing using dideoxynucleotide chain terminators dd. ATP, dd. GTP, dd. CTP, and dd. TTP.

07_01. jpg

07_02. jpg

07_02_2. jpg

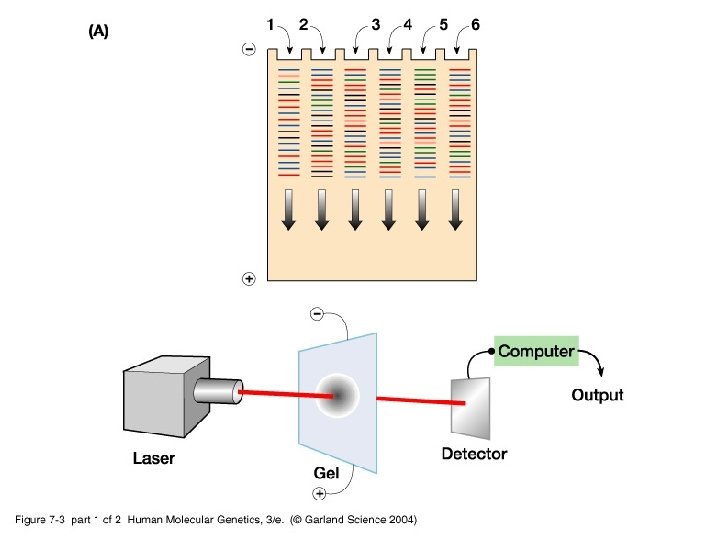
• Automated DNA sequencing using fluorophores and capillary gel electrophoresis.

07_03. jpg

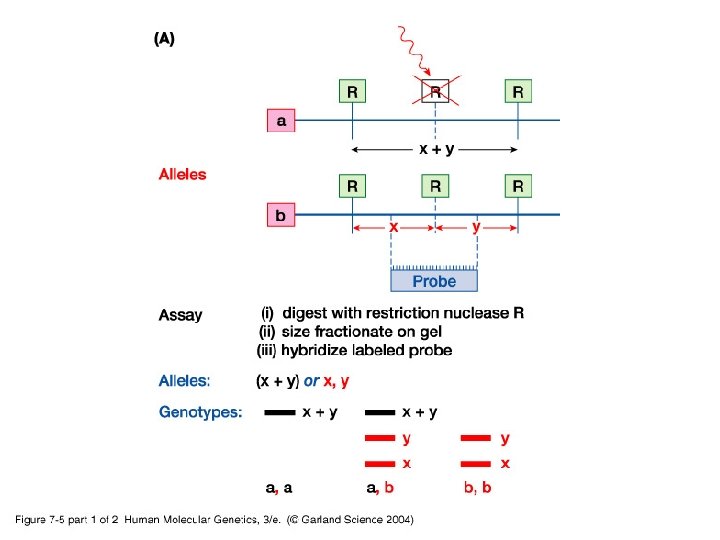
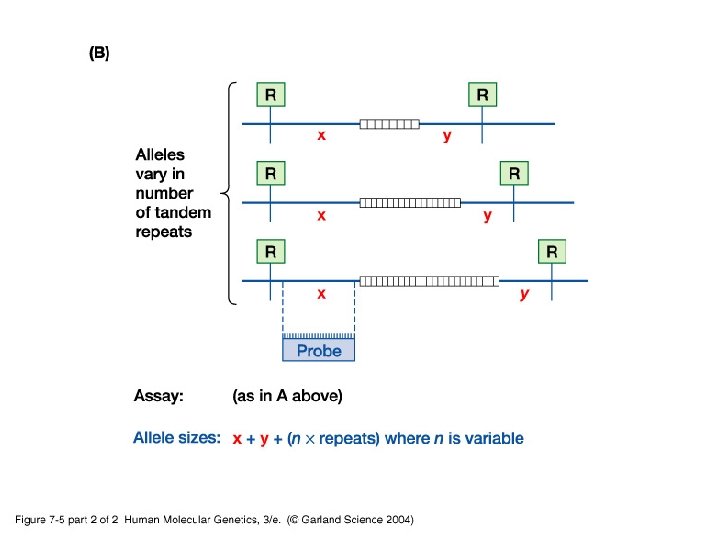
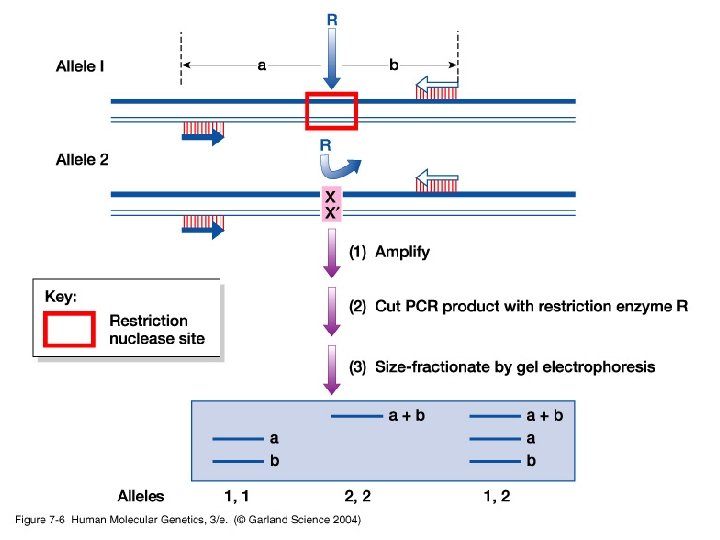
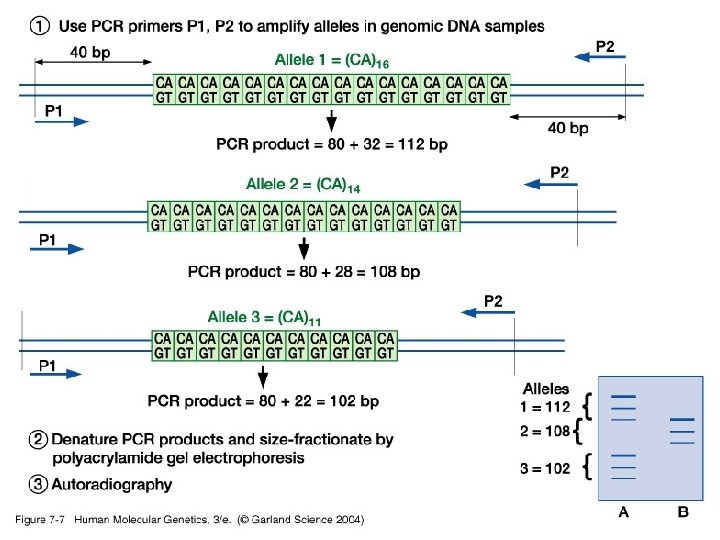
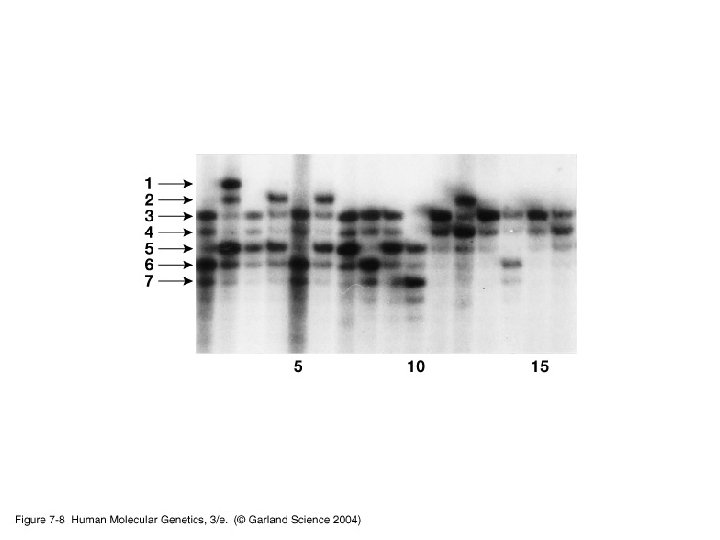
• Simple/basic genotyping by restriction site polymorphisms (RSPs) and varaible number tandem repeats (VNTRs) - RSPs: single nucleotide polymorphism may cause a loss or gain in a restriction site generating an RSP. Used in identifying carriers for some disease causing genes. - VNTR: use of PCR or Southern blot hybridization to identify differences in the number of microsatellite tandem repeats.

07_05. jpg

07_05_2. jpg

07_06. jpg

07_07. jpg

07_08. jpg

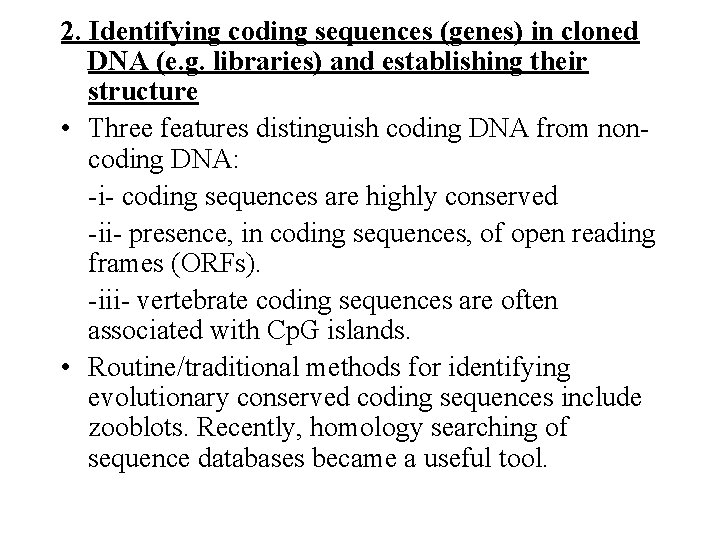
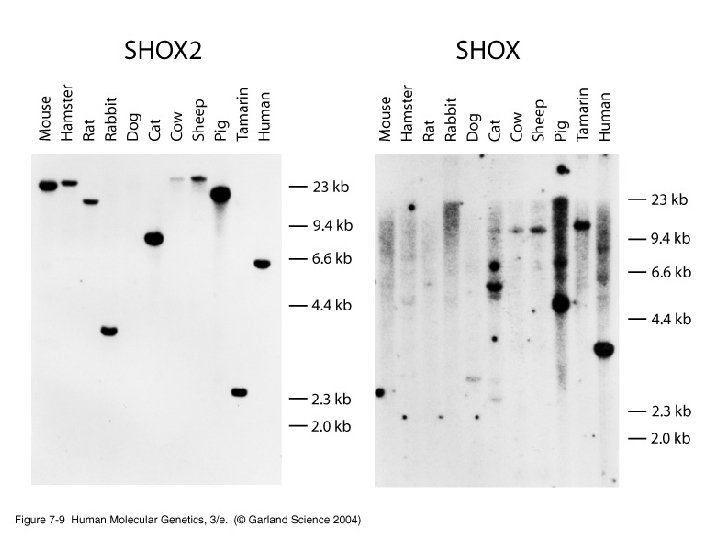
2. Identifying coding sequences (genes) in cloned DNA (e. g. libraries) and establishing their structure • Three features distinguish coding DNA from noncoding DNA: -i- coding sequences are highly conserved -ii- presence, in coding sequences, of open reading frames (ORFs). -iii- vertebrate coding sequences are often associated with Cp. G islands. • Routine/traditional methods for identifying evolutionary conserved coding sequences include zooblots. Recently, homology searching of sequence databases became a useful tool.

07_09. jpg

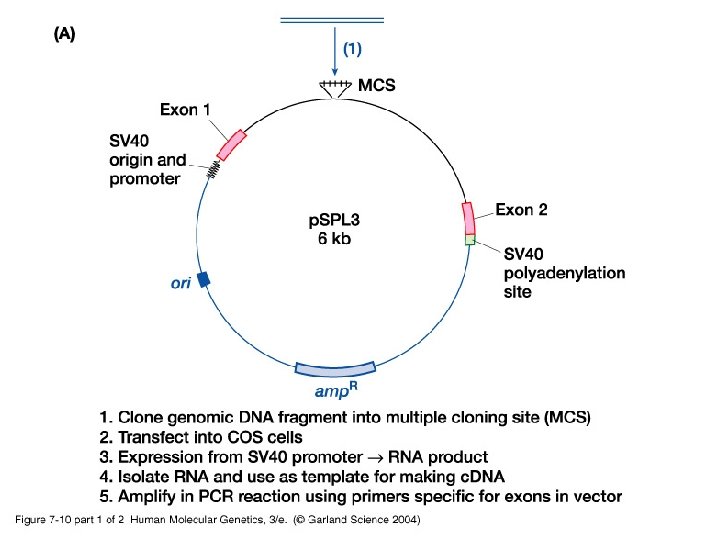
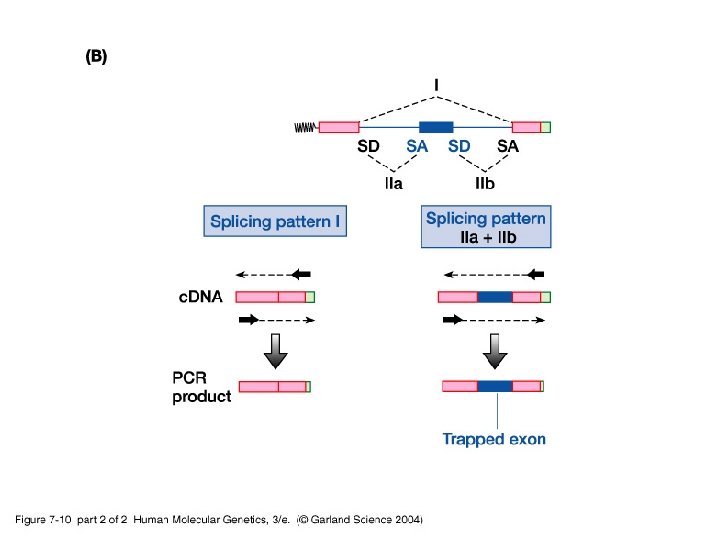
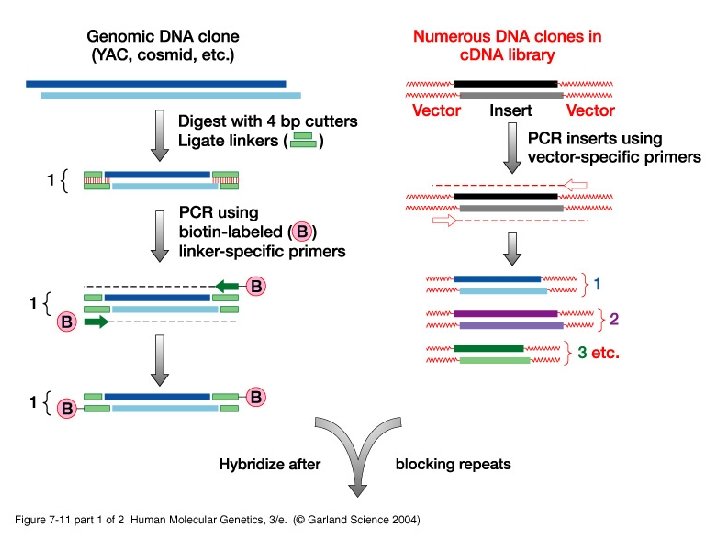
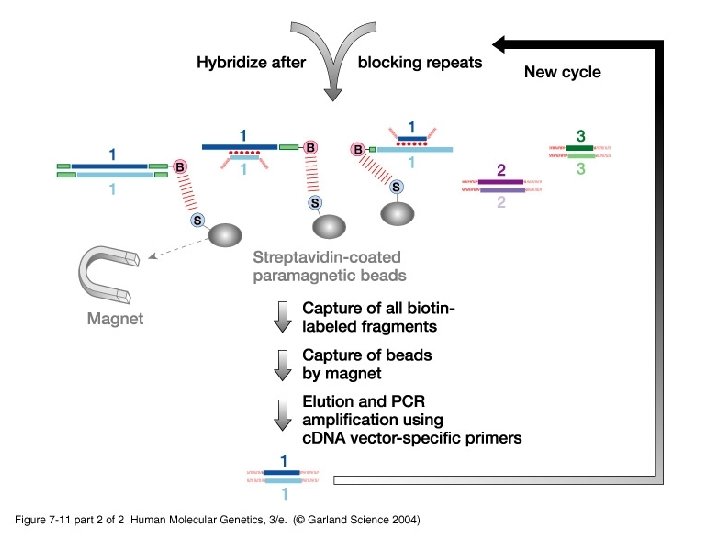
• Besides routine methods, new more specialized procedures are used to identify coding sequences: -i- Exon trapping uses an artificial RNA splicing assay. -ii- c. DNA selection by heteroduplex formation using magnetic beads capture identifies expressed sequences in genomic clones.

07_10. jpg

07_10_2. jpg

07_11. jpg

07_11_2. jpg

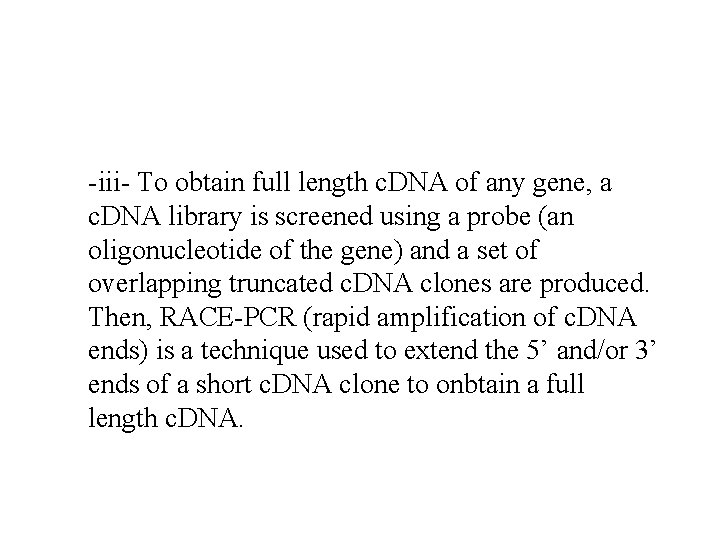
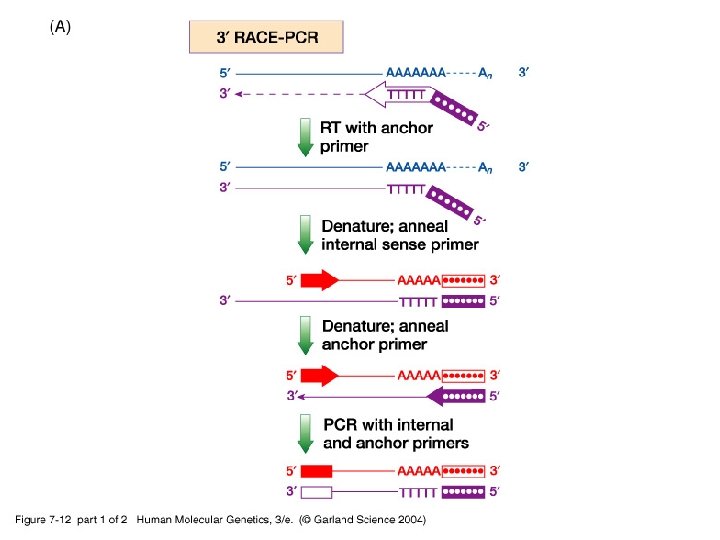
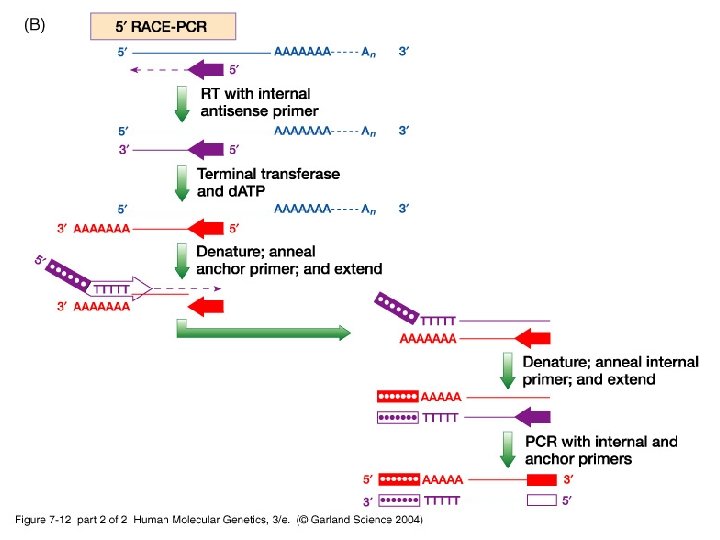
-iii- To obtain full length c. DNA of any gene, a c. DNA library is screened using a probe (an oligonucleotide of the gene) and a set of overlapping truncated c. DNA clones are produced. Then, RACE-PCR (rapid amplification of c. DNA ends) is a technique used to extend the 5’ and/or 3’ ends of a short c. DNA clone to onbtain a full length c. DNA.

07_12. jpg

07_12_2. jpg

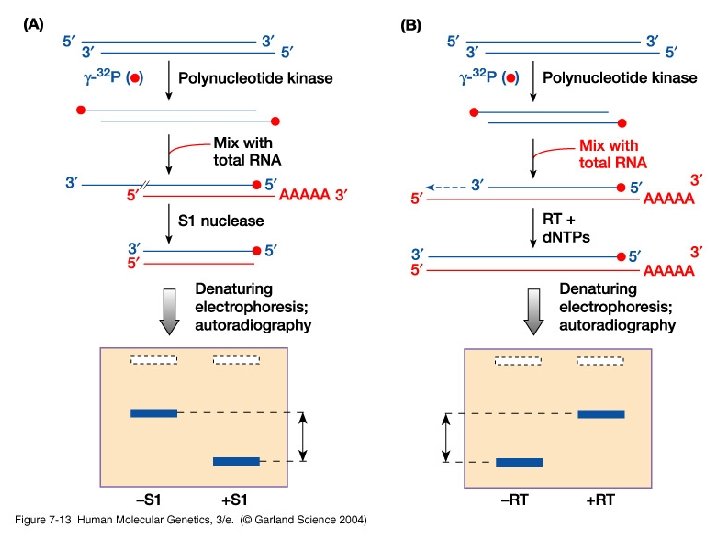
-iv- Mapping transcription start site could be achieved by S 1 nuclease protection or primer extension.

07_13. jpg

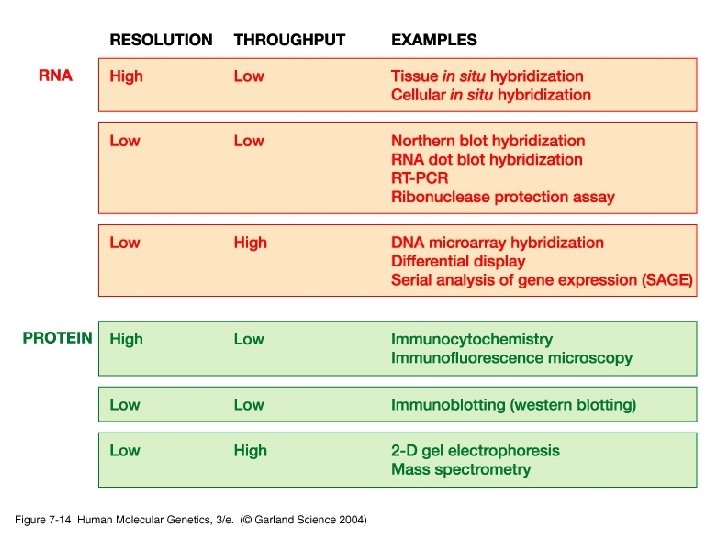
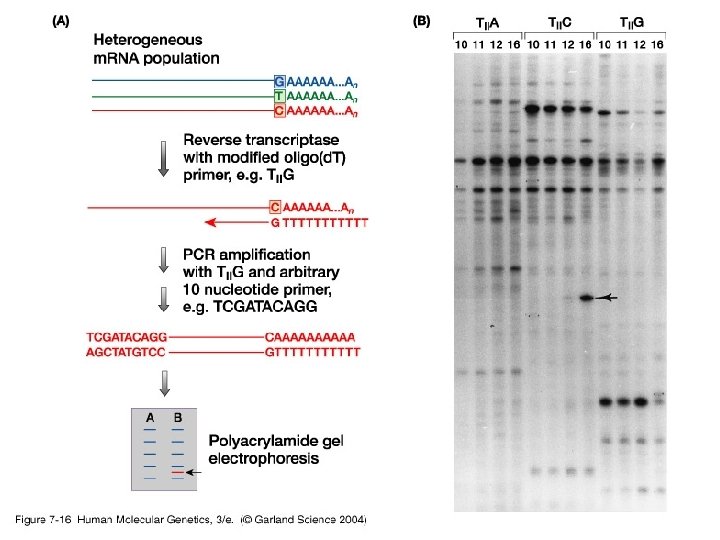
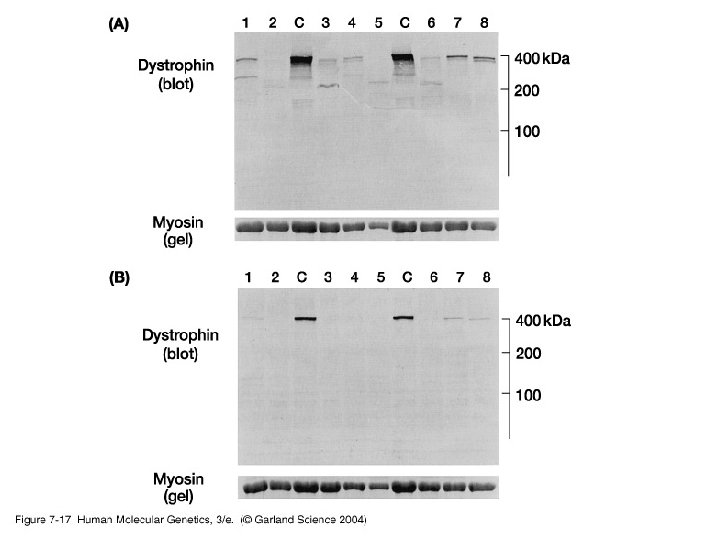
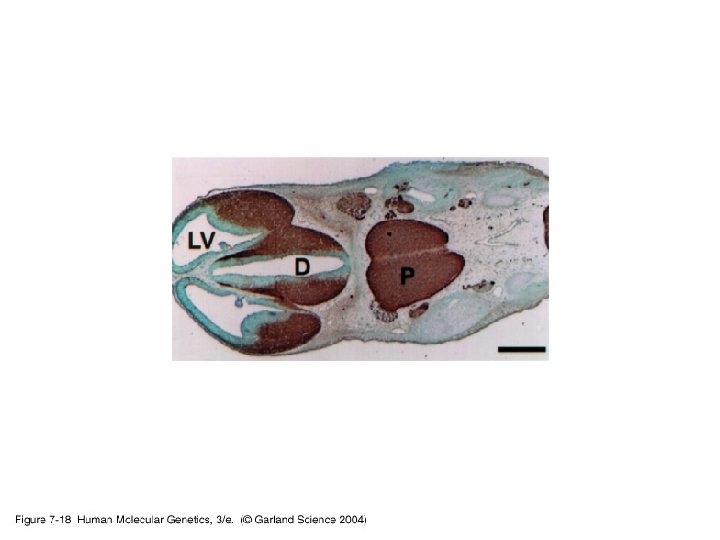
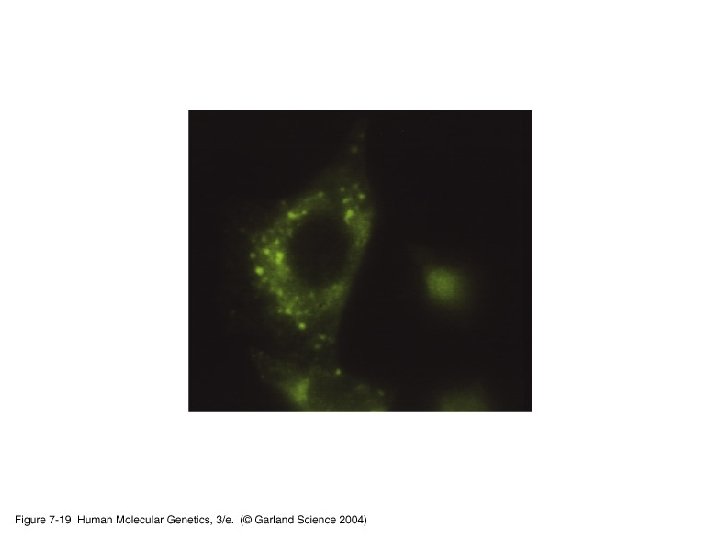
3. Studying gene expression • Principles of expression screening – in vitro versus in vivo. RNA analysis versus tissues and individual cells.

07_14. jpg

07_15. jpg

07_16. jpg

07_17. jpg

07_18. jpg

07_19. jpg