CHAPTER 6 PHYLOGENETIC TREE RECONSTRUCTION INTRODUCTION DISTANCE BASED
CHAPTER 6 PHYLOGENETIC TREE RECONSTRUCTION INTRODUCTION ØDISTANCE BASED TREE ØCHARACTER BASED TREE Ø
INTRODUCTION l l l All life is related by common ancestor Phylogenetic = phylo + genetic phylo fr. Greek phulon meaning race, class genetics= Scientific study of the principles of heredity and the variation of inherited traits among related organisms. Phylogeny = the sequence of events involved in the evolutionary development of a species or taxonomic group of organisms
Traditional Vs. Modern Trees l l The traditional phylogenetic tree using phenotypes (physical characteristics) to infer their genotypes (the genes that give rise to their physical characeristics) If the phenotypes were similar, it was assumed that the genes that coded for the phenotypes were also similar and vice versa.

l l l Originally the phenotypes examined consisted largely of gross anatomic features Later, behavioural, ultrastructural and biochemical characteristics were also studied Basis for many evolutionary studies until today Has some limitations The modern phylogenetic tree using genotypes to infer their phenotypes
Modern Trees- Advantages of molecular data 1. Molecular data less likely to suffer from problems associated with convergent evolution (similar phenotypes can evolve in 2 distantly related organisms) e. g eyes in human, flies and mollusks. Light detecting organ. All have different physical features. Misleading evolutionary relationship. Phenotypic similarities do not always reflects genetic similarities.
2. Not many phenotypic features suitable for comparison. Difficult to study. e. g the study of bacterial taxonomic have always been problematic because bact. Have few obvious traits. Even at microscopic level, few have same traits etc. Molecular data only have 4 DNA bases and the differences among them is less and manageable to study
3. When study 2 distantly related organisms, its difficult to find a most common phenotype features. e. g. if we want to study bacteria, worms and mammals, what are the common phenotypic features should be compared? l Whereas for molecular data only have 4 DNA bases and easy to handle. 4. Analysis that rely on DNA and protein sequences are free from such problems because many homologous molecules are essential to all living things
TERMINOLOGIES l l Phylogeny or evolutionary tree- the actual pattern of historical relationship among taxa Tree – a mathematical structure which is used to model the actual evolutionary history of a group of sequences or organisms A tree consists of nodes connected by branches (also called edges) Terminal nodes/leaves/ terminal taxa – represent sequences or organisms for which we have data, maybe either extant or extinct
l l l Internal nodes – represent hypothetical ancestor Root- the ancestor of ALL the sequences or organisms that comprise the tree Taxon/taxa – species or gene under investigation Ingroup – a collection of taxa/taxon under investigation Outgroup- collection of closely related the whole or partial ingroup, use to root the tree Character – data used to build the tree e. g DNA or protein data

Terminal Nodes (leaf) Internal Nodes Hypothetical ancestor Branch Root/ ancestor
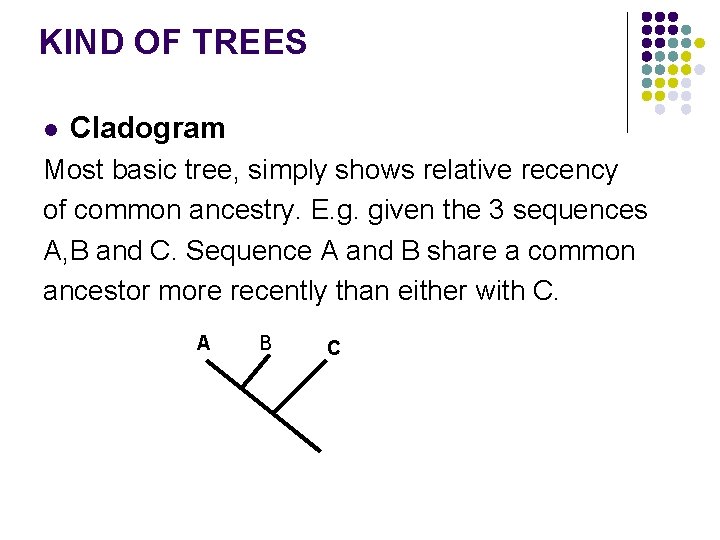
KIND OF TREES l Cladogram Most basic tree, simply shows relative recency of common ancestry. E. g. given the 3 sequences A, B and C. Sequence A and B share a common ancestor more recently than either with C. A B C

Additive Tree Contains additional information, namely branch lengths. l These are numbers associated with each branch that correspond to some attribute of the sequences such as the evolutionary change. l Other name include ‘phylogram’ and ‘metric tree’. l
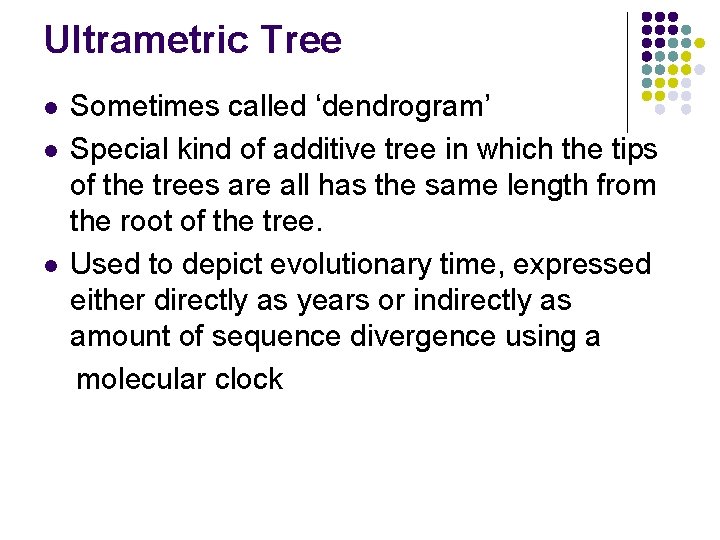
Ultrametric Tree l l l Sometimes called ‘dendrogram’ Special kind of additive tree in which the tips of the trees are all has the same length from the root of the tree. Used to depict evolutionary time, expressed either directly as years or indirectly as amount of sequence divergence using a molecular clock
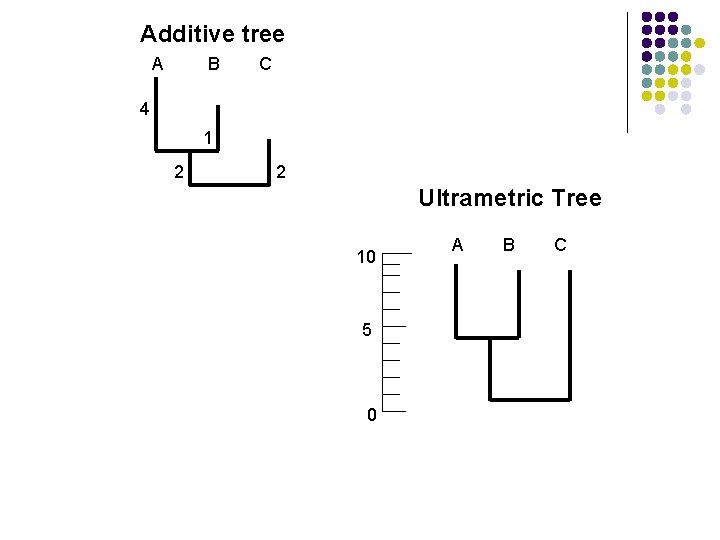
Additive tree A B C 4 1 2 2 Ultrametric Tree 10 5 0 A B C
ROOTED TREE Has a node identified as the root from which the ultimately all other nodes descend. l Hence, the rooted tree has a direction that correspond to the evolutionary time. l The closer a node is to the root of the tree, the older it is in time and vice versa l Allows us to define ancestor-descendent relationship l

UNROOTED TREE l l l Lacks a root and hence, do not specify the evolutionary relationship among its ancestor -descendent Say nothing about the direction in which the evolution occurred Sequences that maybe adjacent on this tree need not be evolutionarily closely related
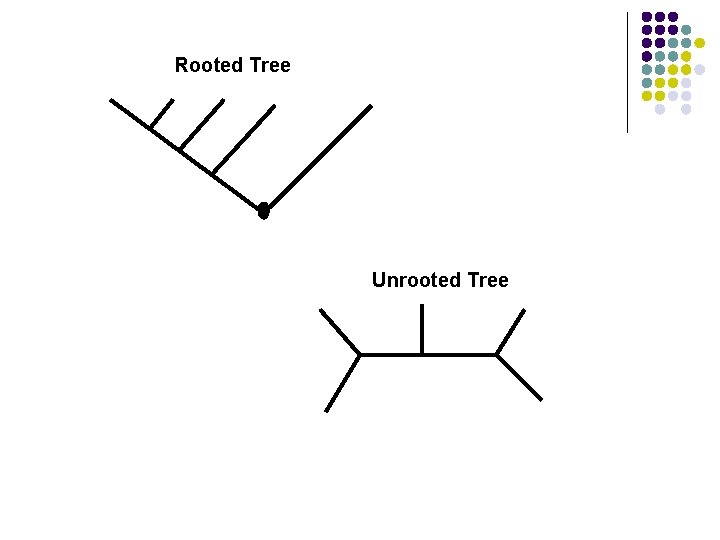
Rooted Tree Unrooted Tree
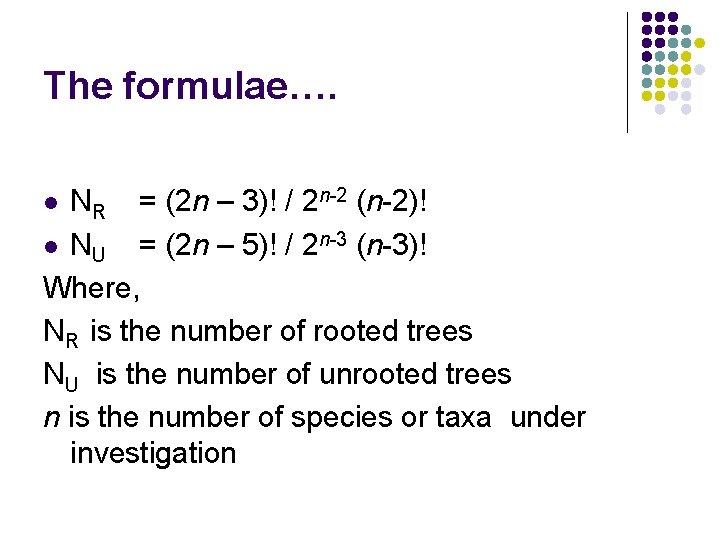
The formulae…. NR = (2 n – 3)! / 2 n-2 (n-2)! l NU = (2 n – 5)! / 2 n-3 (n-3)! Where, NR is the number of rooted trees NU is the number of unrooted trees n is the number of species or taxa under investigation l
Description of the formulae l l Despite the so many of the possible rooted and unrooted trees can be drawn from the formula, only ONE possible tree can represent the true phylogenetic relationship among the genes or species being considered Hence, most phylogenetic trees generated with molecular data are refferred to as inferred trees
- Slides: 19