Chapter 6 Molecular Signatures of Natural Selection ChauTi
























- Slides: 83

Chapter 6 Molecular Signatures of Natural Selection Chau-Ti Ting ctting@ntu. edu. tw Unless noted, the course materials are licensed under Creative Commons Attribution-Non. Commercial-Share. Alike 3. 0 Taiwan (CC BY-NC-SA 3. 0) 1

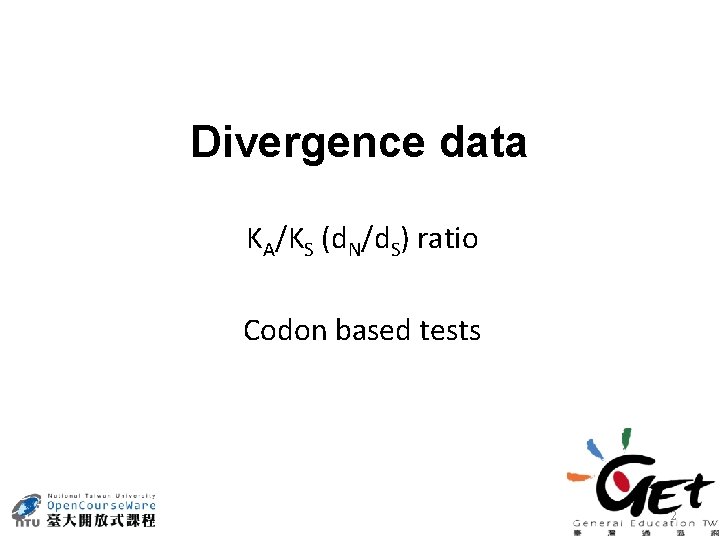
Divergence data KA/KS (d. N/d. S) ratio Codon based tests 2

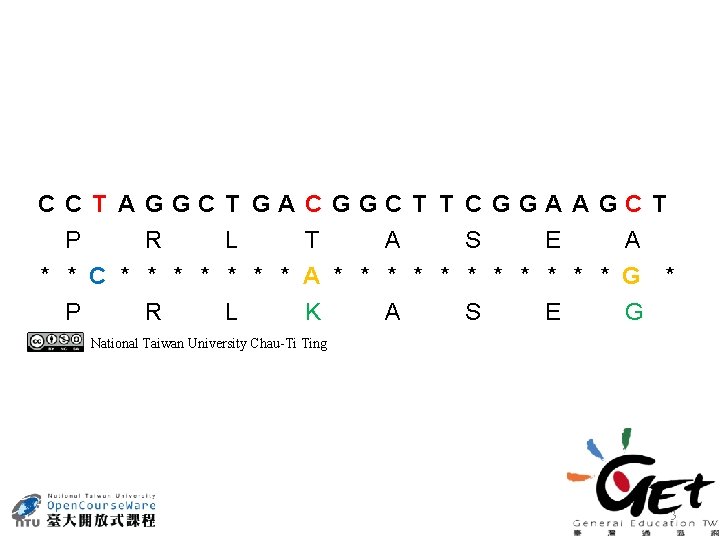
CCT AGGCT GACGGCT T CGGAAGCT P R L T A S E A * * C * * * * A * * * G * P R L K A S E G National Taiwan University Chau-Ti Ting 3

CCT AGGCT GACGGCT T CGGAAGCT P R L T A S E A * * C * * * * A * * * G * P R L K A S E G National Taiwan University Chau-Ti Ting 4

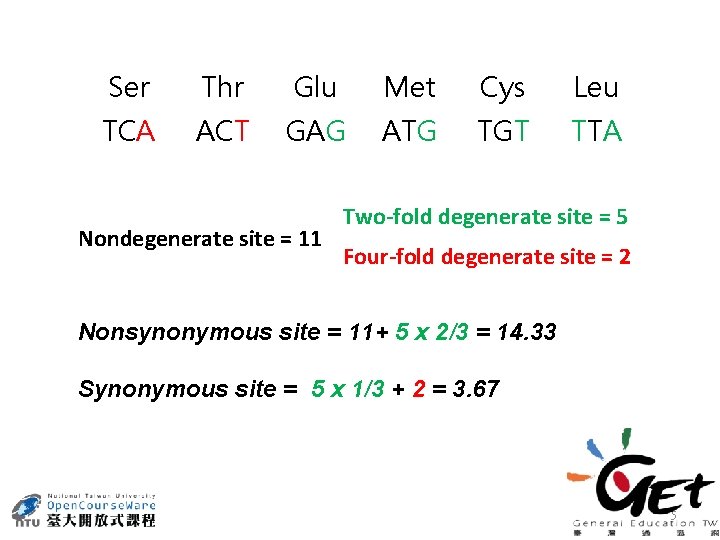
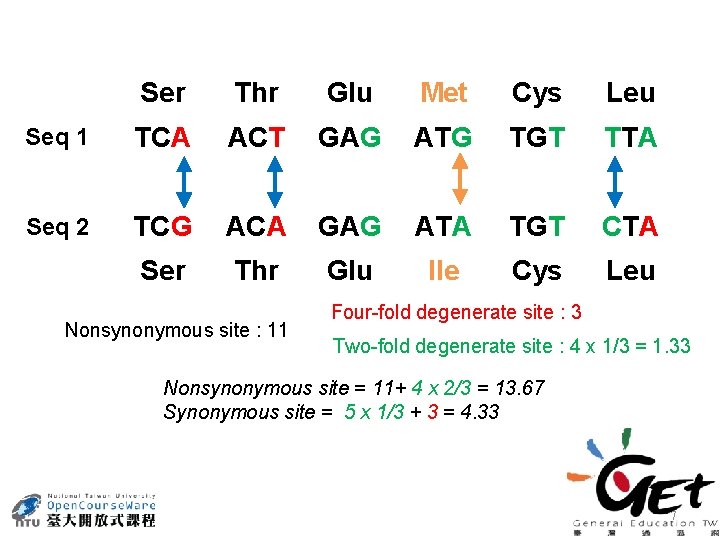
Ser Thr Glu Met Cys Leu TCA ACT GAG ATG TGT TTA Nondegenerate site = 11 Two-fold degenerate site = 5 Four-fold degenerate site = 2 Nonsynonymous site = 11+ 5 x 2/3 = 14. 33 Synonymous site = 5 x 1/3 + 2 = 3. 67 5

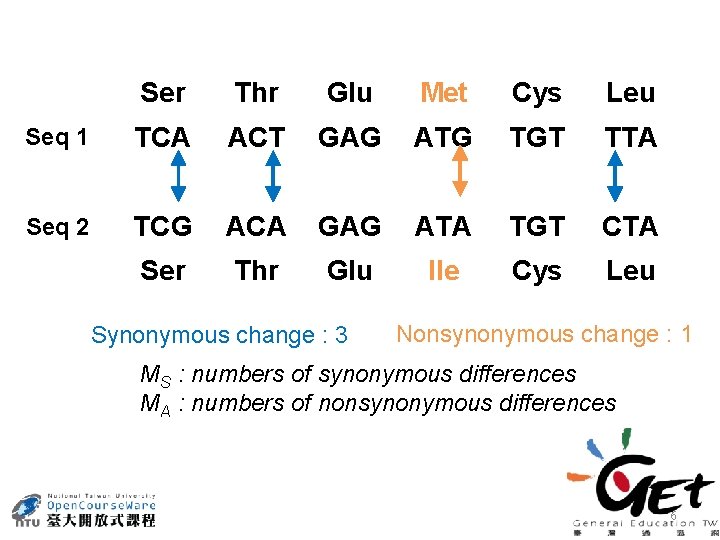
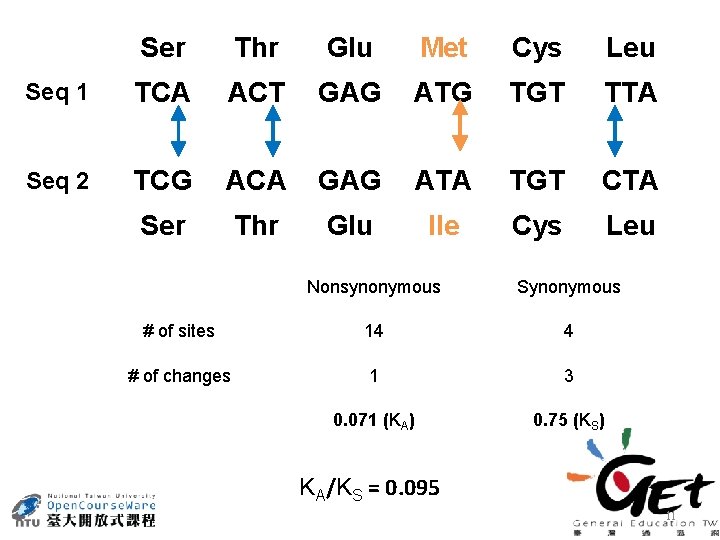
Ser Thr Glu Met Cys Leu Seq 1 TCA ACT GAG ATG TGT TTA Seq 2 TCG ACA GAG ATA TGT CTA Ser Thr Glu Ile Cys Leu Synonymous change : 3 Nonsynonymous change : 1 MS : numbers of synonymous differences MA : numbers of nonsynonymous differences 6

Ser Thr Glu Met Cys Leu Seq 1 TCA ACT GAG ATG TGT TTA Seq 2 TCG ACA GAG ATA TGT CTA Ser Thr Glu Ile Cys Leu Nonsynonymous site : 11 Four-fold degenerate site : 3 Two-fold degenerate site : 4 x 1/3 = 1. 33 Nonsynonymous site = 11+ 4 x 2/3 = 13. 67 Synonymous site = 5 x 1/3 + 3 = 4. 33 7

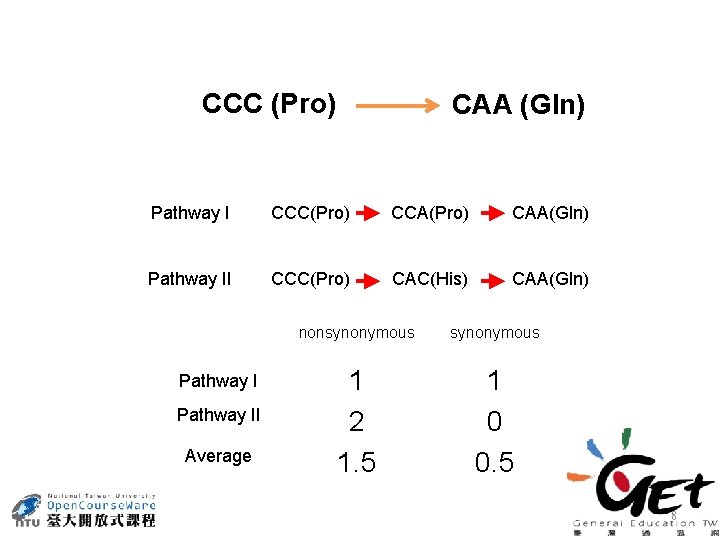
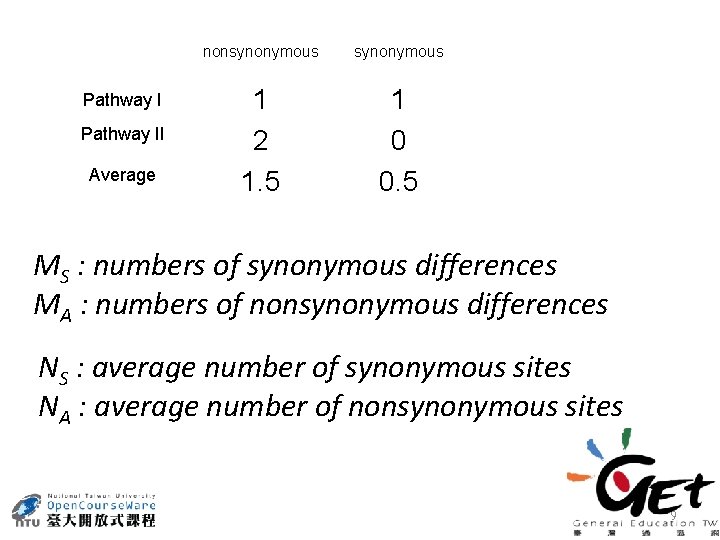
CCC (Pro) CAA (Gln) Pathway I CCC(Pro) CCA(Pro) CAA(Gln) Pathway II CCC(Pro) CAC(His) CAA(Gln) Pathway II Average nonsynonymous 1 2 1. 5 1 0 0. 5 8

Pathway II Average nonsynonymous 1 2 1. 5 1 0 0. 5 MS : numbers of synonymous differences MA : numbers of nonsynonymous differences NS : average number of synonymous sites NA : average number of nonsynonymous sites 9

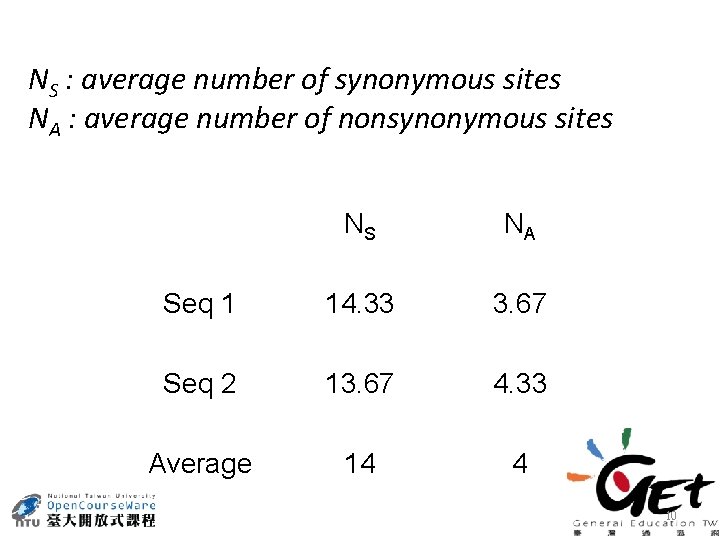
NS : average number of synonymous sites NA : average number of nonsynonymous sites NS NA Seq 1 14. 33 3. 67 Seq 2 13. 67 4. 33 Average 14 4 10

Ser Thr Glu Met Cys Leu Seq 1 TCA ACT GAG ATG TGT TTA Seq 2 TCG ACA GAG ATA TGT CTA Ser Thr Glu Ile Cys Leu Nonsynonymous Synonymous # of sites 14 4 # of changes 1 3 0. 071 (KA) 0. 75 (KS) KA/KS = 0. 095 11

12

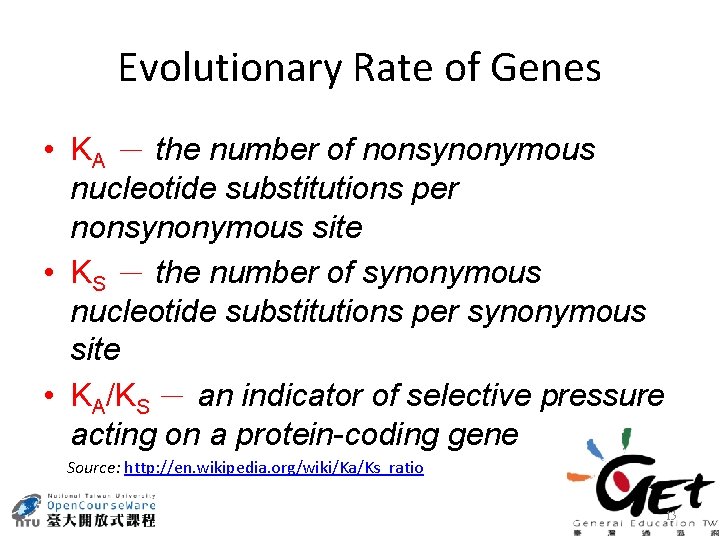
Evolutionary Rate of Genes • KA - the number of nonsynonymous nucleotide substitutions per nonsynonymous site • KS - the number of synonymous nucleotide substitutions per synonymous site • KA/KS - an indicator of selective pressure acting on a protein-coding gene Source: http: //en. wikipedia. org/wiki/Ka/Ks_ratio 13

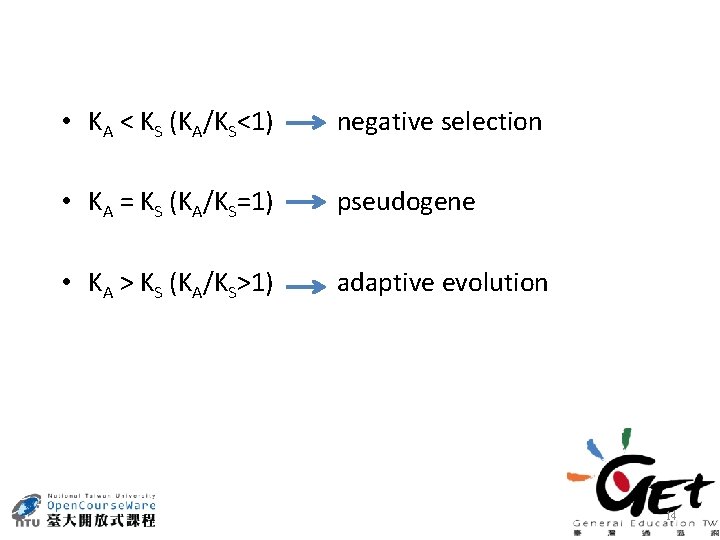
• KA < KS (KA/KS<1) negative selection • KA = KS (KA/KS=1) pseudogene • KA > KS (KA/KS>1) adaptive evolution 14

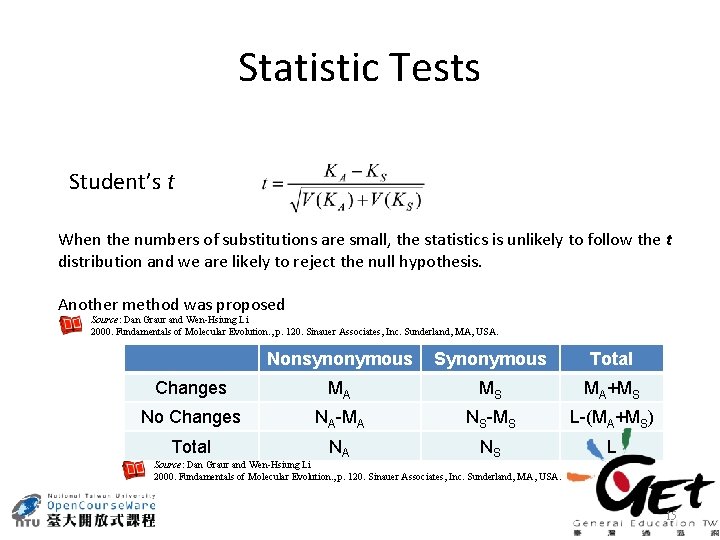
Statistic Tests Student’s t When the numbers of substitutions are small, the statistics is unlikely to follow the t distribution and we are likely to reject the null hypothesis. Another method was proposed Source: Dan Graur and Wen-Hsiung Li 2000. Fundamentals of Molecular Evolution. , p. 120. Sinauer Associates, Inc. Sunderland, MA, USA. Nonsynonymous Synonymous Total Changes MA MS MA+MS No Changes NA-MA NS-MS L-(MA+MS) Total NA NS L Source: Dan Graur and Wen-Hsiung Li 2000. Fundamentals of Molecular Evolution. , p. 120. Sinauer Associates, Inc. Sunderland, MA, USA. 15

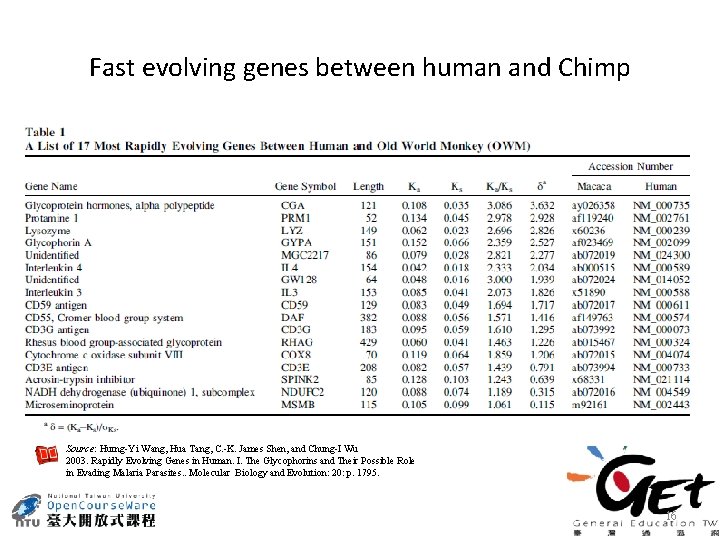
Fast evolving genes between human and Chimp Source: Hurng-Yi Wang, Hua Tang, C. -K. James Shen, and Chung-I Wu 2003. Rapidly Evolving Genes in Human. I. The Glycophorins and Their Possible Role in Evading Malaria Parasites. . Molecular Biology and Evolution: 20: p. 1795. 16

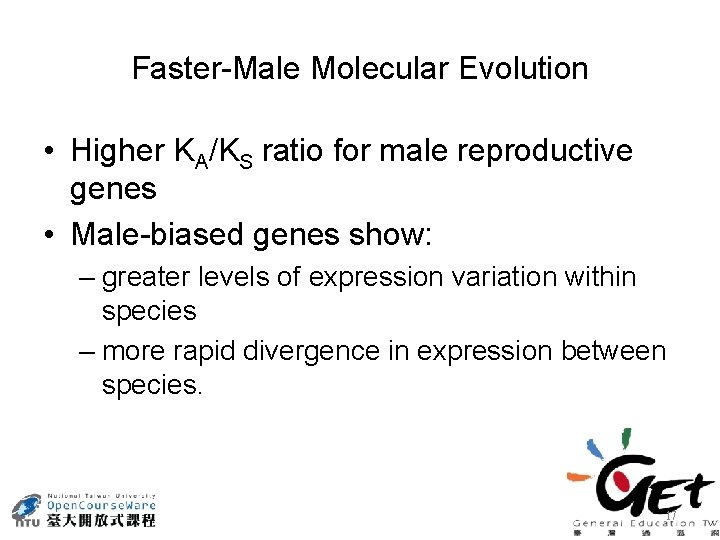
Faster-Male Molecular Evolution • Higher KA/KS ratio for male reproductive genes • Male-biased genes show: – greater levels of expression variation within species – more rapid divergence in expression between species. 17

Source: Willie J. Swanson, Andrew G. Clark, Heidi M. Waldrip-Dail, Mariana F. Wolfner, and Charles F. Aquadro 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. . PNAS 98: p. 7375 Fig. 2. The number of nonsynonymous substitutions per nonsynonymous site (d. N) plotted against the number of synonymous substitutions per synonymous site (d. S) for the D. simulans accessory gland EST library (A), and random nonreproductive proteins (B) compared with D. melanogaster genomic sequence. are putative Acps (by differential hybridization or containing signal sequences), whereas cannot be identified as putative Acps by these methods. The line shows the neutral expectation of d. N = d. S. Data for the lower panel are from Moriyama and Powell (44). d. N and d. S were estimated by the maximum likelihood method (50, 51). Similar results are obtained by using the method of Nei and Gojobori (53). (Swanson et al, 2001) Source: Willie J. Swanson, Andrew G. Clark, Heidi M. Waldrip-Dail, Mariana F. Wolfner, and Charles F. Aquadro 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. . PNAS 98: p. 7375 18

Codon based test (PAML) Source: http: //abacus. gene. ucl. ac. uk/software/paml. html Mol Biol Evol (2007) 24 (8): 1586 -1591. doi: 10. 1093/molbev/msm 088 19

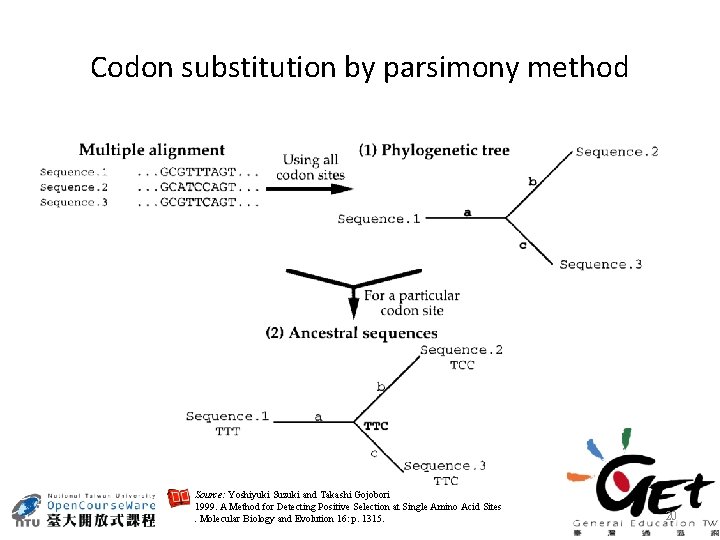
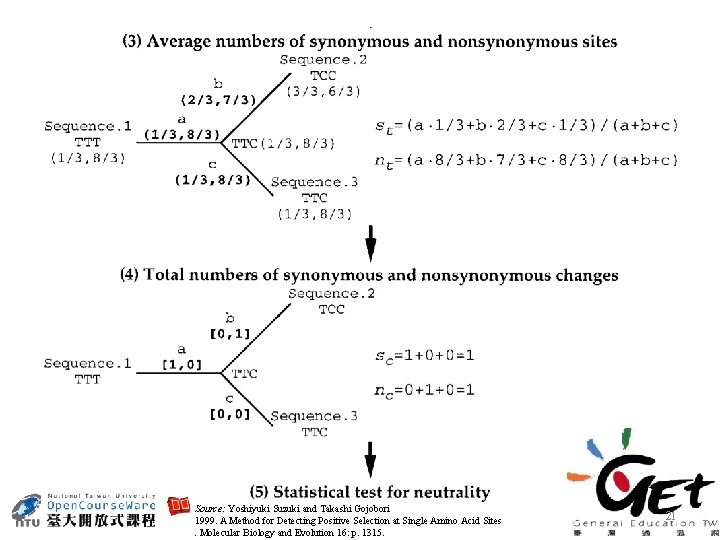
Codon substitution by parsimony method Source: Yoshiyuki Suzuki and Takashi Gojobori 1999. A Method for Detecting Positive Selection at Single Amino Acid Sites. Molecular Biology and Evolution 16: p. 1315. 20

Source: Yoshiyuki Suzuki and Takashi Gojobori 1999. A Method for Detecting Positive Selection at Single Amino Acid Sites. Molecular Biology and Evolution 16: p. 1315. 21

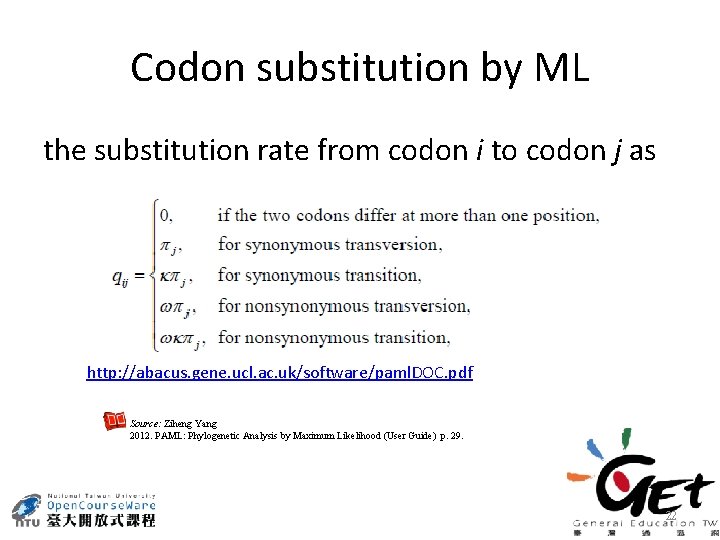
Codon substitution by ML the substitution rate from codon i to codon j as http: //abacus. gene. ucl. ac. uk/software/paml. DOC. pdf Source: Ziheng Yang 2012. PAML: Phylogenetic Analysis by Maximum Likelihood (User Guide) p. 29. 22

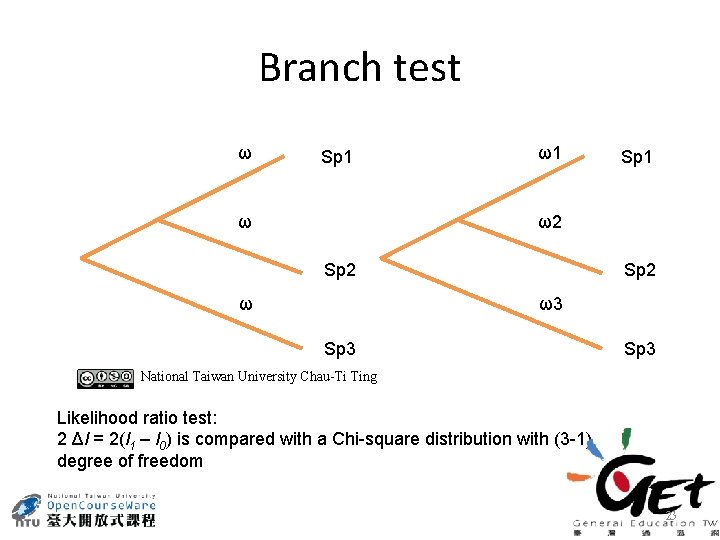
Branch test ω Sp 1 ω ω1 Sp 1 ω2 Sp 2 ω3 Sp 3 National Taiwan University Chau-Ti Ting Likelihood ratio test: 2 Δl = 2(l 1 – l 0) is compared with a Chi-square distribution with (3 -1) degree of freedom 23

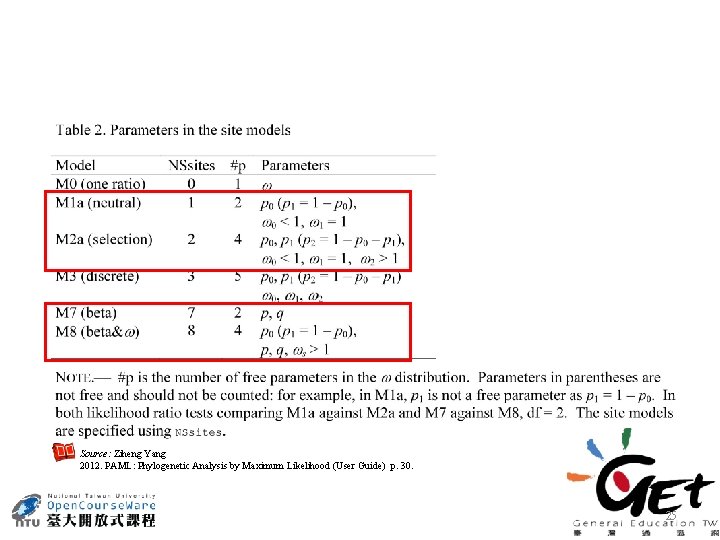
Site test • Assuming ω follows discrete-gamma distribution, positive selection is tested using a likelihood ratio test comparing a null model that does not allow ω > 1 with an alternative model that does. • Assuming ω follows beta distribution, positive selection is tested using a likelihood ratio test comparing a null model that does not allow ω > 1 with an alternative model that does. Source: Ziheng Yang 2006. Computational Molecular Evolution, p. 274 -275. Oxford University Press. , London, UK. 24

Source: Ziheng Yang 2012. PAML: Phylogenetic Analysis by Maximum Likelihood (User Guide) p. 30. 25

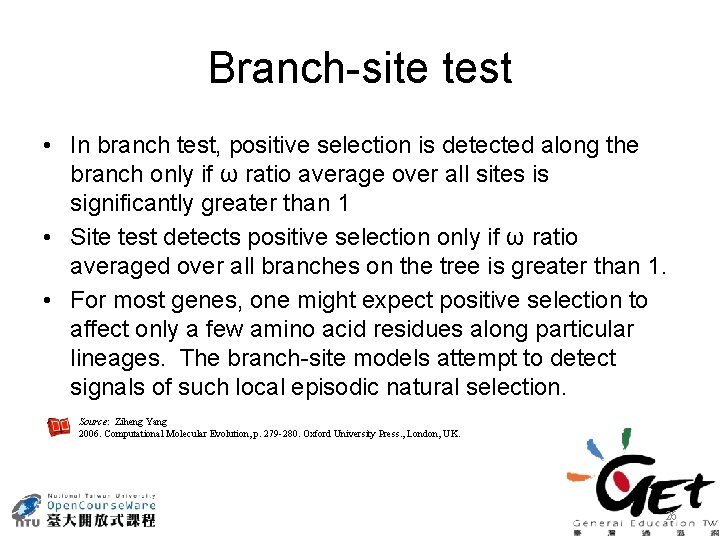
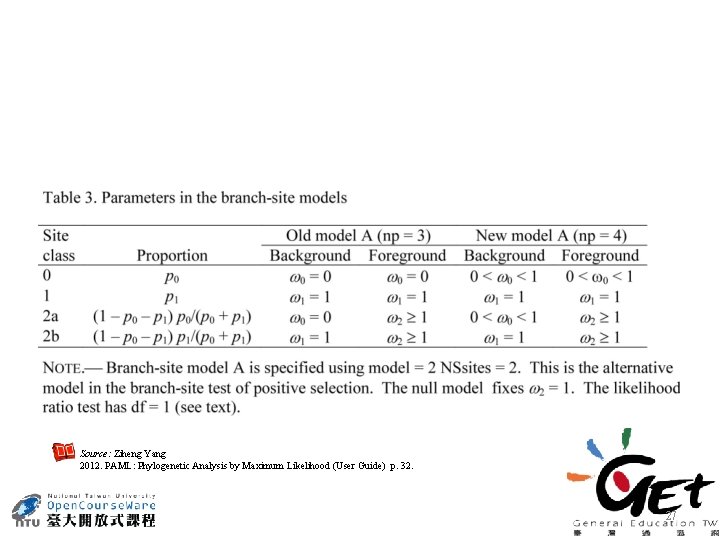
Branch-site test • In branch test, positive selection is detected along the branch only if ω ratio average over all sites is significantly greater than 1 • Site test detects positive selection only if ω ratio averaged over all branches on the tree is greater than 1. • For most genes, one might expect positive selection to affect only a few amino acid residues along particular lineages. The branch-site models attempt to detect signals of such local episodic natural selection. Source: Ziheng Yang 2006. Computational Molecular Evolution, p. 279 -280. Oxford University Press. , London, UK. 26

Source: Ziheng Yang 2012. PAML: Phylogenetic Analysis by Maximum Likelihood (User Guide) p. 32. 27

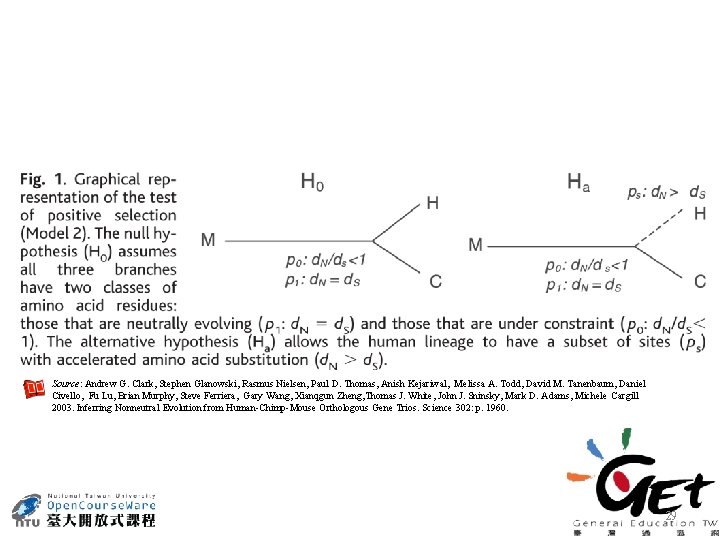
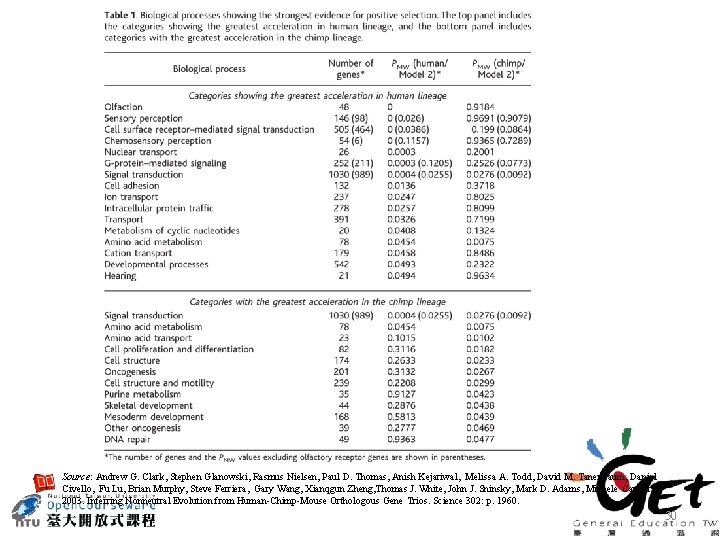
Inferring Nonneutral Evolution from Human. Chimp-Mouse Orthologous Gene Trios Science. 2003. Vol. 302. pp 1960 -1963. http: //www. sciencemag. org/content/302/5652/1960. abstract 28

Source: Andrew G. Clark, Stephen Glanowski, Rasmus Nielsen, Paul D. Thomas, Anish Kejariwal, Melissa A. Todd, David M. Tanenbaum, Daniel Civello, Fu Lu, Brian Murphy, Steve Ferriera, Gary Wang, Xianqgun Zheng, Thomas J. White, John J. Sninsky, Mark D. Adams, Michele Cargill 2003. Inferring Nonneutral Evolution from Human-Chimp-Mouse Orthologous Gene Trios. Science 302: p. 1960. 29

Source: Andrew G. Clark, Stephen Glanowski, Rasmus Nielsen, Paul D. Thomas, Anish Kejariwal, Melissa A. Todd, David M. Tanenbaum, Daniel Civello, Fu Lu, Brian Murphy, Steve Ferriera, Gary Wang, Xianqgun Zheng, Thomas J. White, John J. Sninsky, Mark D. Adams, Michele Cargill 2003. Inferring Nonneutral Evolution from Human-Chimp-Mouse Orthologous Gene Trios. Science 302: p. 1960. 30

Source: Andrew G. Clark, Stephen Glanowski, Rasmus Nielsen, Paul D. Thomas, Anish Kejariwal, Melissa A. Todd, David M. Tanenbaum, Daniel Civello, Fu Lu, Brian Murphy, Steve Ferriera, Gary Wang, Xianqgun Zheng, Thomas J. White, John J. Sninsky, Mark D. Adams, Michele Cargill 2003. Inferring Nonneutral Evolution from Human-Chimp-Mouse Orthologous Gene Trios. Science 302: p. 1960. 31

Possible pitfall • Mouse is only distant-related to chimpanzee and human – may cause problems to infer directionality of changes • Using more closely-related outgroup such as macaque may help to improve such difficulty 32

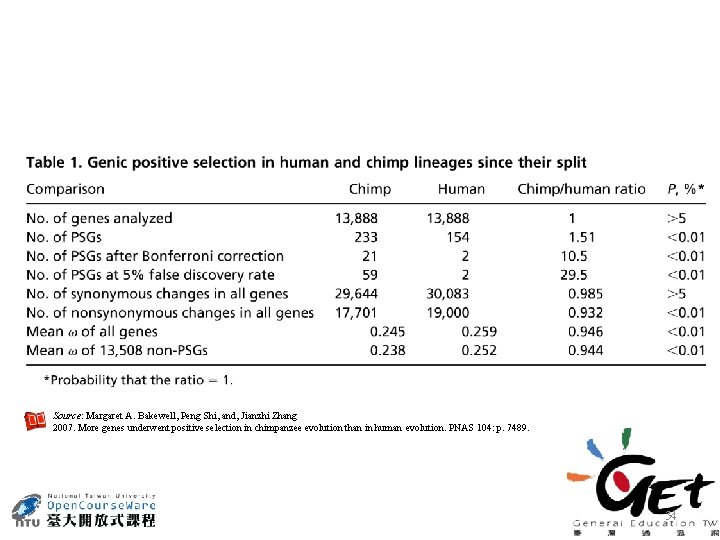
More genes underwent positive selection in chimpanzee evolution than in human evolution PNAS. 2007. Vol. 104. pp 7489 -7494. http: //www. pnas. org/content/104/18/7489 33

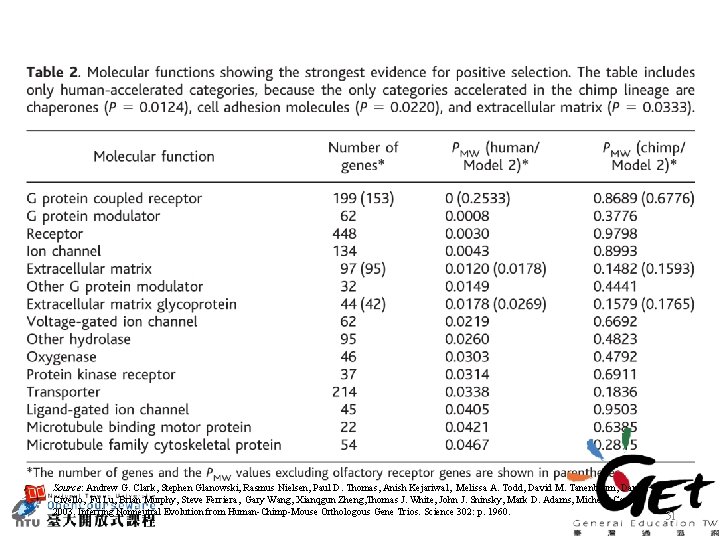
Source: Margaret A. Bakewell, Peng Shi, and, Jianzhi Zhang 2007. More genes underwent positive selection in chimpanzee evolution than in human evolution. PNAS 104: p. 7489. 34

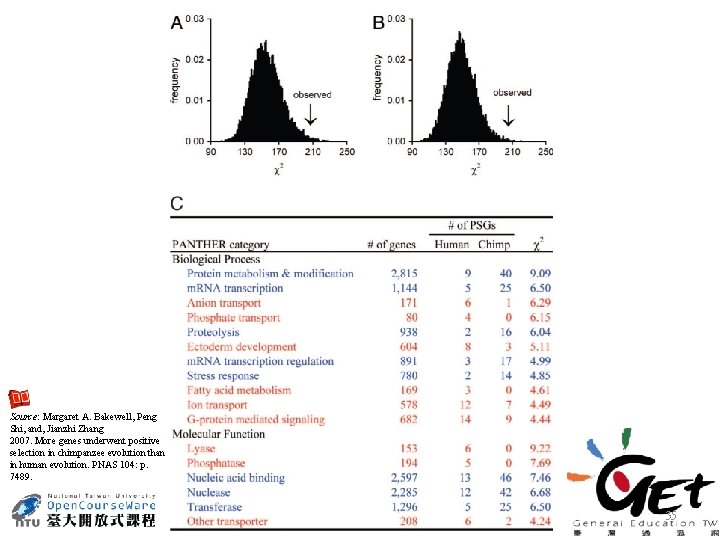
Source: Margaret A. Bakewell, Peng Shi, and, Jianzhi Zhang 2007. More genes underwent positive selection in chimpanzee evolution than in human evolution. PNAS 104: p. 7489. 35

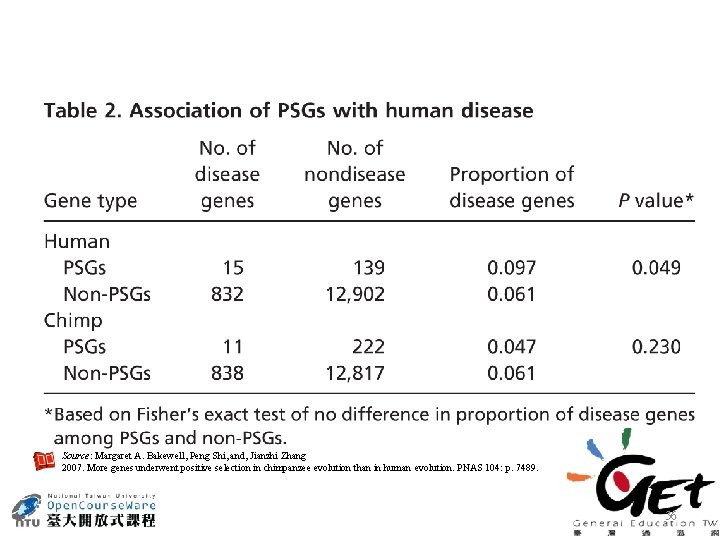
Source: Margaret A. Bakewell, Peng Shi, and, Jianzhi Zhang 2007. More genes underwent positive selection in chimpanzee evolution than in human evolution. PNAS 104: p. 7489. 36

Population Based Methods: Comparing divergence with polymorphism 37

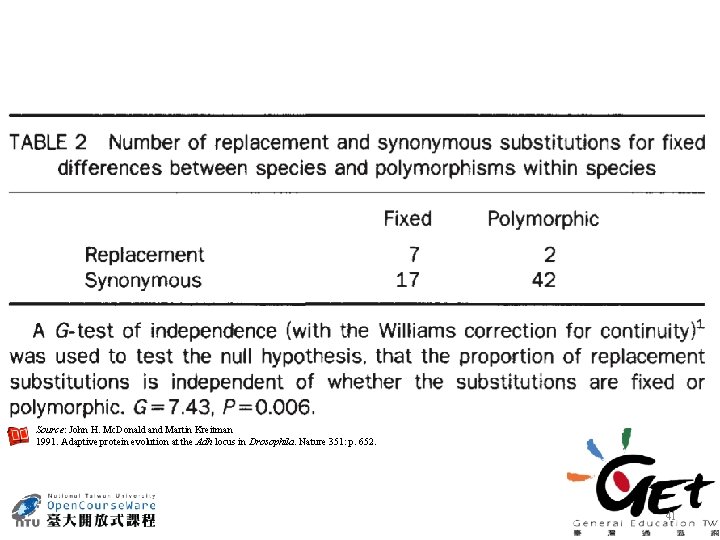
Mc. Donald-Kreitman Test Adaptive protein evolution at the Adh locus in Drosophila Nature. 1991. Vol. 351. pp 652 -654. http: //www. nature. com/nature/journal/v 351/n 6328/pdf/351652 a 0. pdf 38

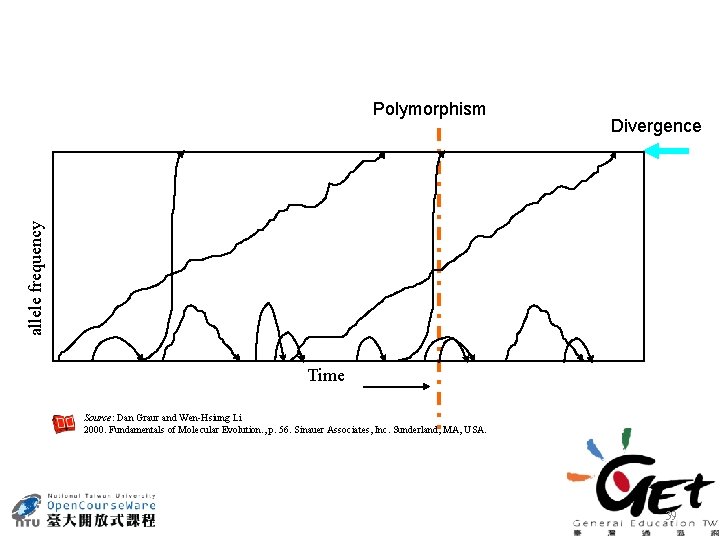
Divergence allele frequency Polymorphism Time Source: Dan Graur and Wen-Hsiung Li 2000. Fundamentals of Molecular Evolution. , p. 56. Sinauer Associates, Inc. Sunderland, MA, USA. 39

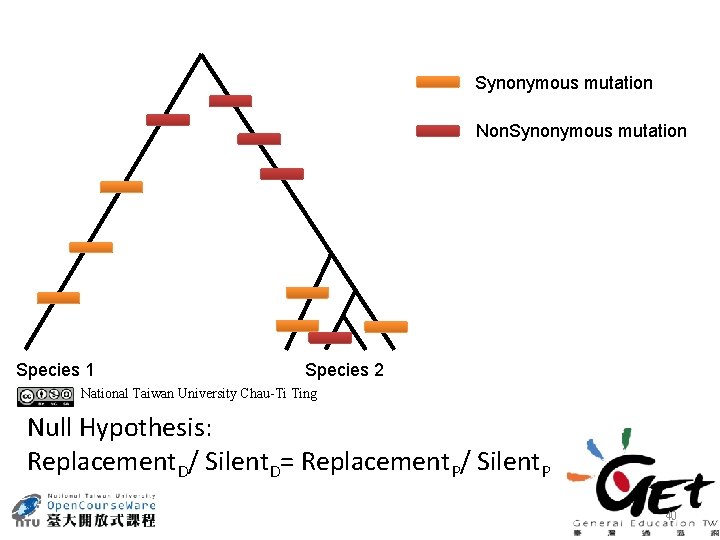
Synonymous mutation Non. Synonymous mutation Species 1 Species 2 National Taiwan University Chau-Ti Ting Null Hypothesis: Replacement. D/ Silent. D= Replacement. P/ Silent. P 40

Source: John H. Mc. Donald and Martin Kreitman 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: p. 652. 41

Frequency spectrum tests (rely on polymorphism data) 42

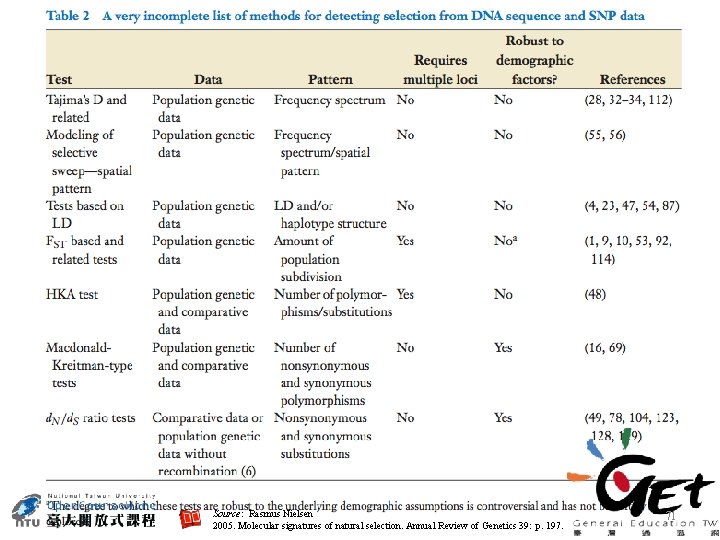
The Frequency Spectrum Source: Rasmus Nielsen 2005. Molecular signatures of natural selection. Annual Review of Genetics 39: p. 197. 43

• Low frequency – – – • Intermediate/high frequency – – • deleterious neutral advantageous Divergence – – neutral advantageous Source: Rasmus Nielsen 2005. Molecular signatures of natural selection. Annual Review of Genetics 39: p. 197. 44

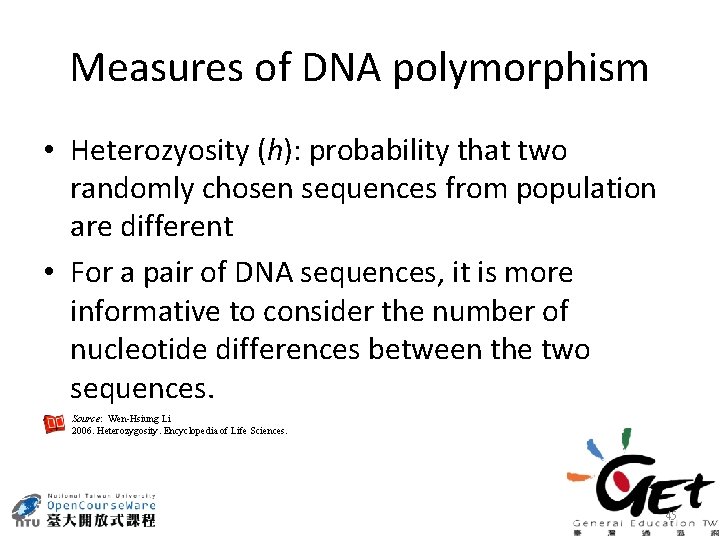
Measures of DNA polymorphism • Heterozyosity (h): probability that two randomly chosen sequences from population are different • For a pair of DNA sequences, it is more informative to consider the number of nucleotide differences between the two sequences. Source: Wen-Hsiung Li 2006. Heterozygosity. Encyclopedia of Life Sciences. 45

National Taiwan University Chau-Ti Ting 46

National Taiwan University Chau-Ti Ting 7 Number of Mutation 6 5 4 3 2 1 0 1 2 National Taiwan University Chau-Ti Ting 3 4 5 6 Ocurrence 47

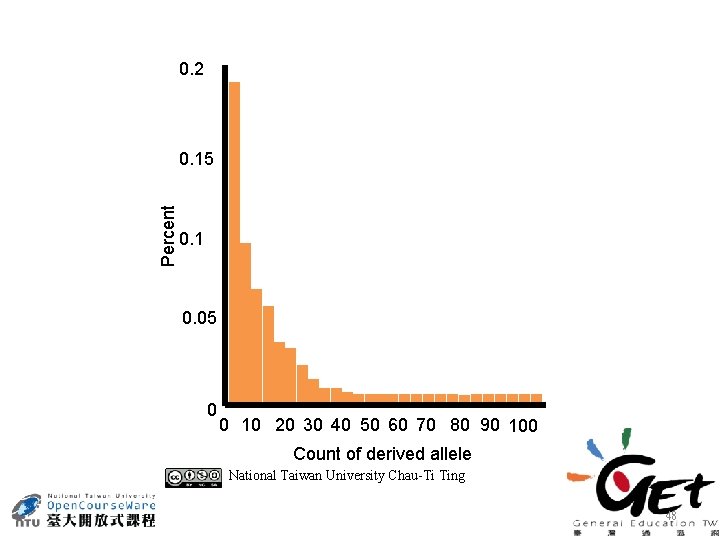
0. 2 Percent 0. 15 0. 1 0. 05 0 0 10 20 30 40 50 60 70 80 90 100 Count of derived allele National Taiwan University Chau-Ti Ting 48

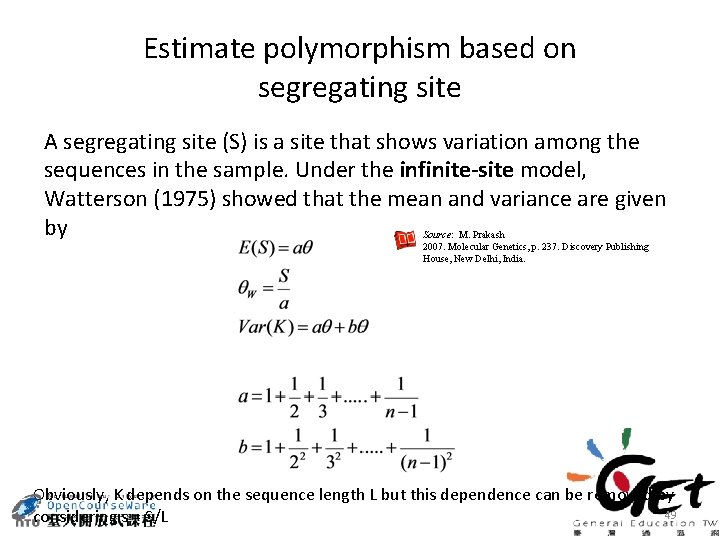
Estimate polymorphism based on segregating site A segregating site (S) is a site that shows variation among the sequences in the sample. Under the infinite-site model, Watterson (1975) showed that the mean and variance are given by Source: M. Prakash 2007. Molecular Genetics, p. 237. Discovery Publishing House, New Delhi, India. Obviously, K depends on the sequence length L but this dependence can be removed by 49 considering s = S/L

Estimate polymorphism based on Nucleotide diversity Π, the average number of nucleotide differences between two sequences randomly chosen from the population Πij is the number of nucleotide differences between the ith and jth sequences and n(n-1)/2 is the number of possible pairs Source: M. Prakash 2007. Molecular Genetics, p. 236. Discovery Publishing House, New Delhi, India 50

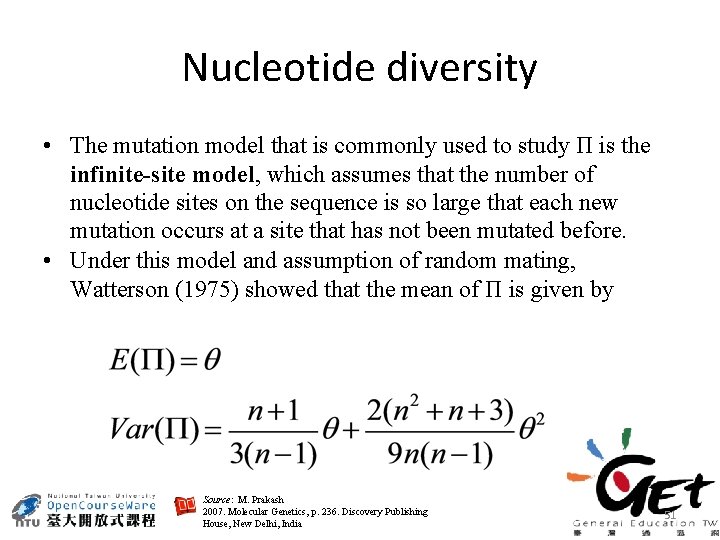
Nucleotide diversity • The mutation model that is commonly used to study Π is the infinite-site model, which assumes that the number of nucleotide sites on the sequence is so large that each new mutation occurs at a site that has not been mutated before. • Under this model and assumption of random mating, Watterson (1975) showed that the mean of Π is given by Source: M. Prakash 2007. Molecular Genetics, p. 236. Discovery Publishing House, New Delhi, India 51

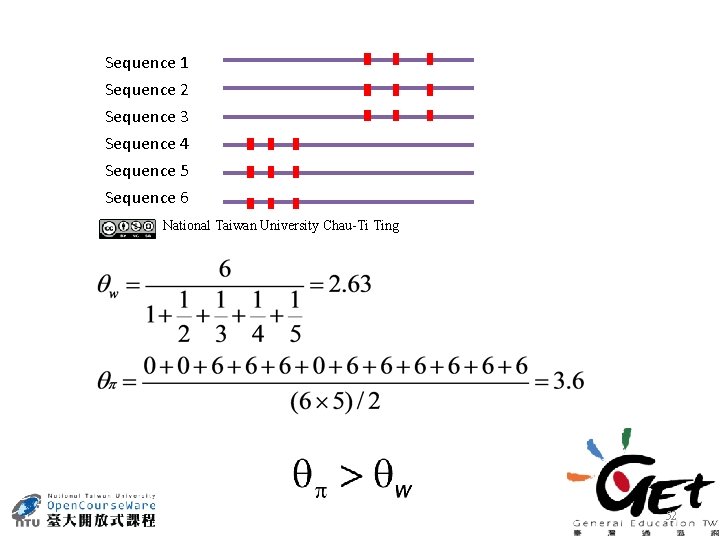
Sequence 1 Sequence 2 Sequence 3 Sequence 4 Sequence 5 Sequence 6 National Taiwan University Chau-Ti Ting 52

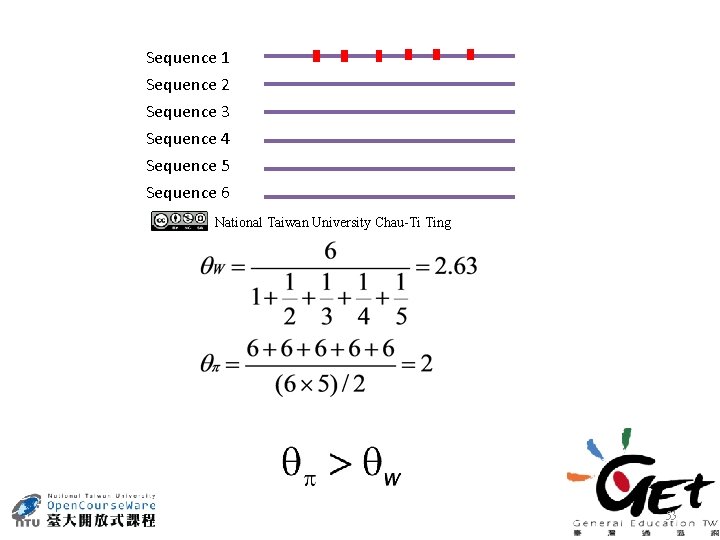
Sequence 1 Sequence 2 Sequence 3 Sequence 4 Sequence 5 Sequence 6 National Taiwan University Chau-Ti Ting 53

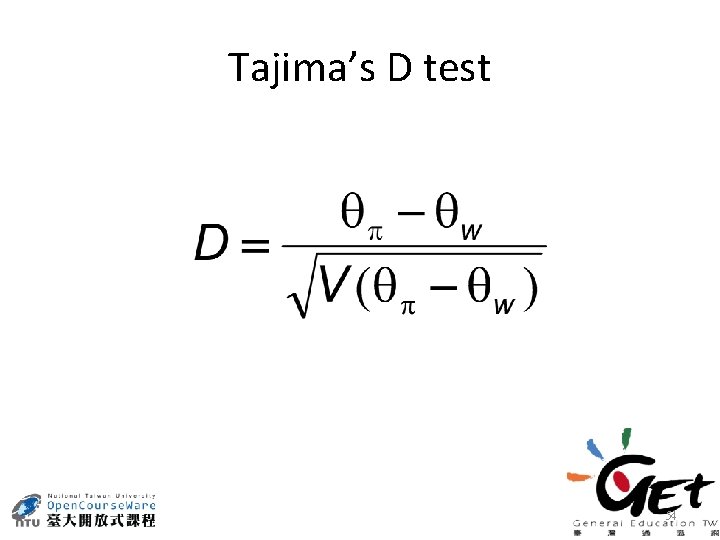
Tajima’s D test 54

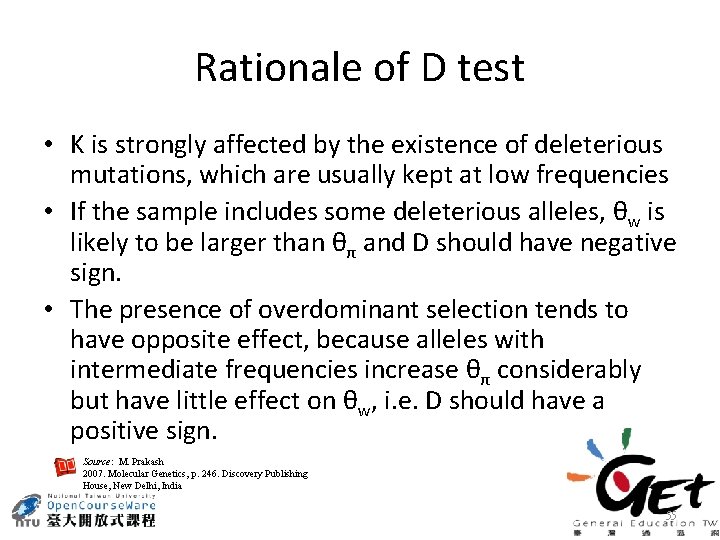
Rationale of D test • K is strongly affected by the existence of deleterious mutations, which are usually kept at low frequencies • If the sample includes some deleterious alleles, θw is likely to be larger than θπ and D should have negative sign. • The presence of overdominant selection tends to have opposite effect, because alleles with intermediate frequencies increase θπ considerably but have little effect on θw, i. e. D should have a positive sign. Source: M. Prakash 2007. Molecular Genetics, p. 246. Discovery Publishing House, New Delhi, India 55

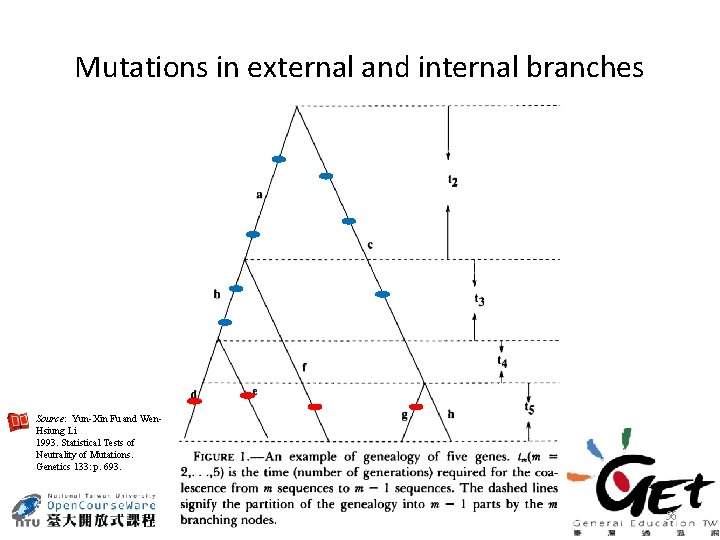
Mutations in external and internal branches Source: Yun-Xin Fu and Wen. Hsiung Li 1993. Statistical Tests of Neutrality of Mutations. Genetics 133: p. 693. 56

57

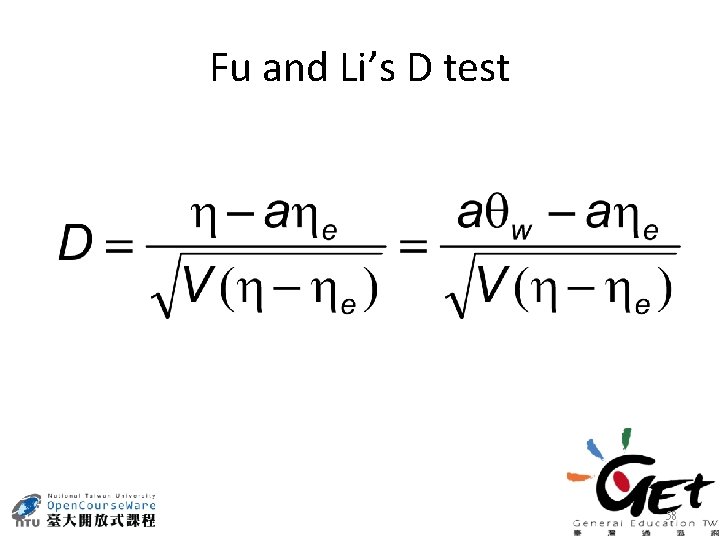
Fu and Li’s D test 58

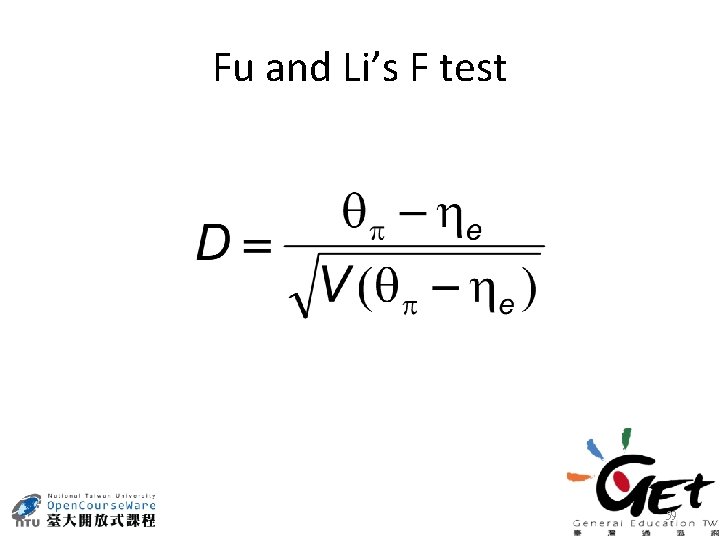
Fu and Li’s F test 59

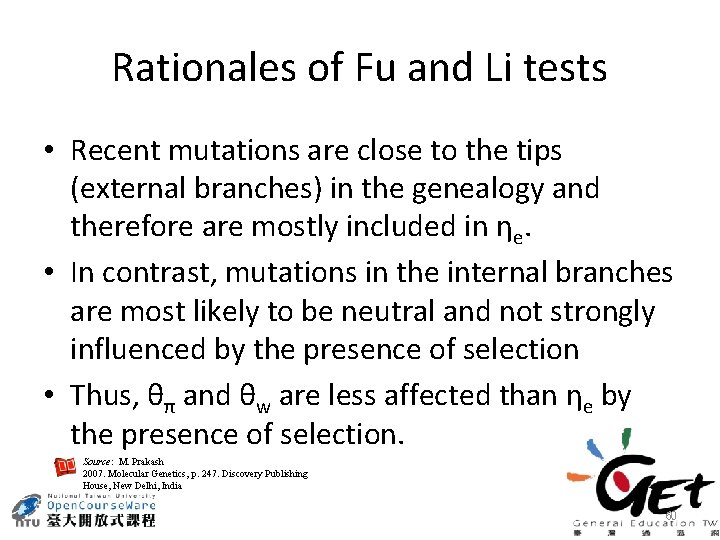
Rationales of Fu and Li tests • Recent mutations are close to the tips (external branches) in the genealogy and therefore are mostly included in ηe. • In contrast, mutations in the internal branches are most likely to be neutral and not strongly influenced by the presence of selection • Thus, θπ and θw are less affected than ηe by the presence of selection. Source: M. Prakash 2007. Molecular Genetics, p. 247. Discovery Publishing House, New Delhi, India 60

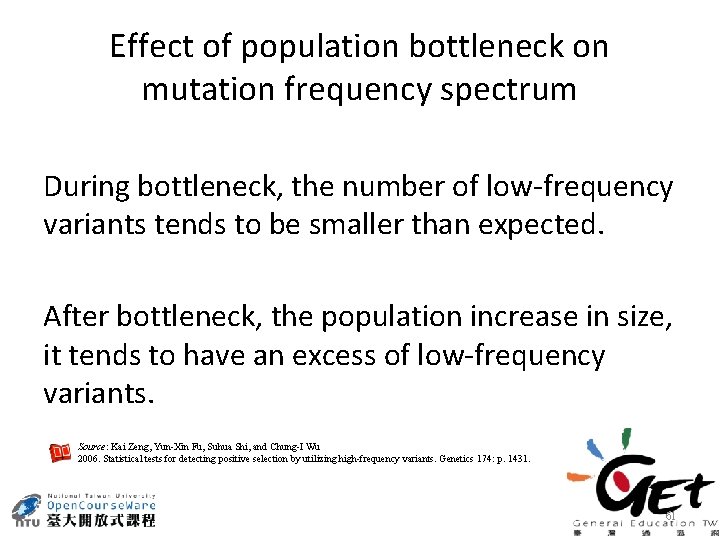
Effect of population bottleneck on mutation frequency spectrum During bottleneck, the number of low-frequency variants tends to be smaller than expected. After bottleneck, the population increase in size, it tends to have an excess of low-frequency variants. Source: Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics 174: p. 1431. 61

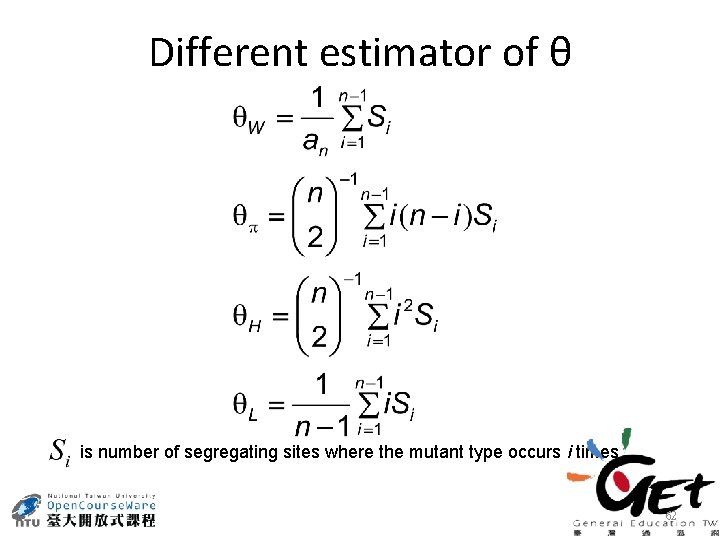
Different estimator of θ is number of segregating sites where the mutant type occurs i times 62

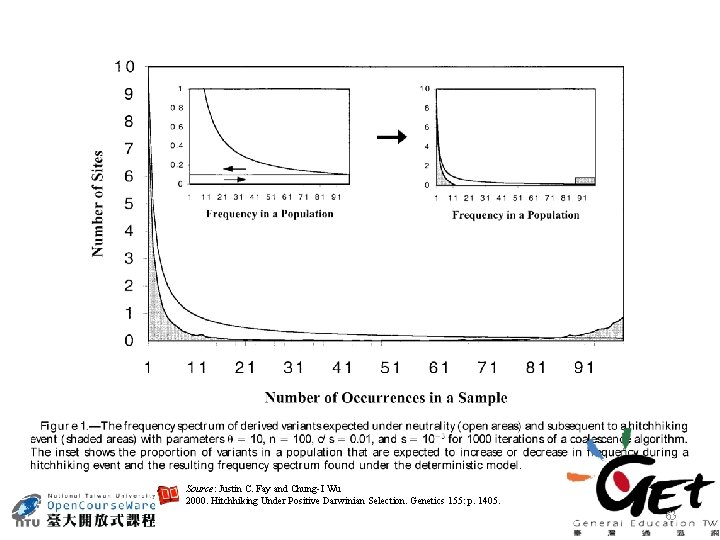
Source: Justin C. Fay and Chung-I Wu 2000. Hitchhiking Under Positive Darwinian Selection. Genetics 155: p. 1405. 63

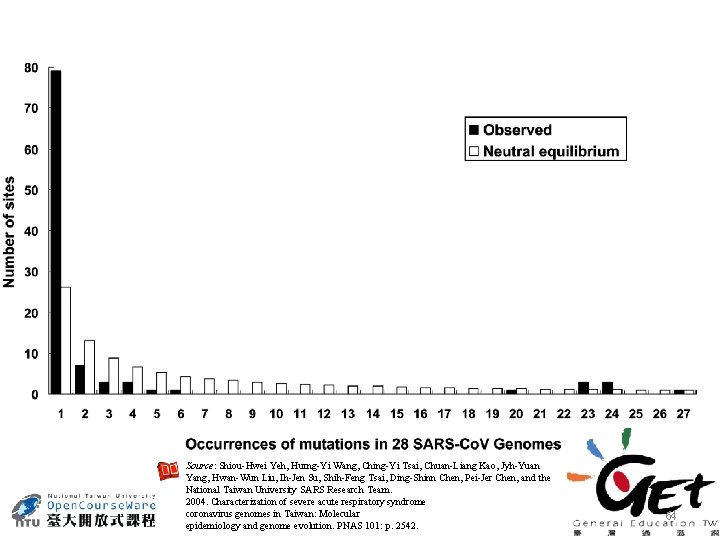
Source: Shiou-Hwei Yeh, Hurng-Yi Wang, Ching-Yi Tsai, Chuan-Liang Kao, Jyh-Yuan Yang, Hwan-Wun Liu, Ih-Jen Su, Shih-Feng Tsai, Ding-Shinn Chen, Pei-Jer Chen, and the National Taiwan University SARS Research Team. 2004. Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: Molecular epidemiology and genome evolution. PNAS 101: p. 2542. 64

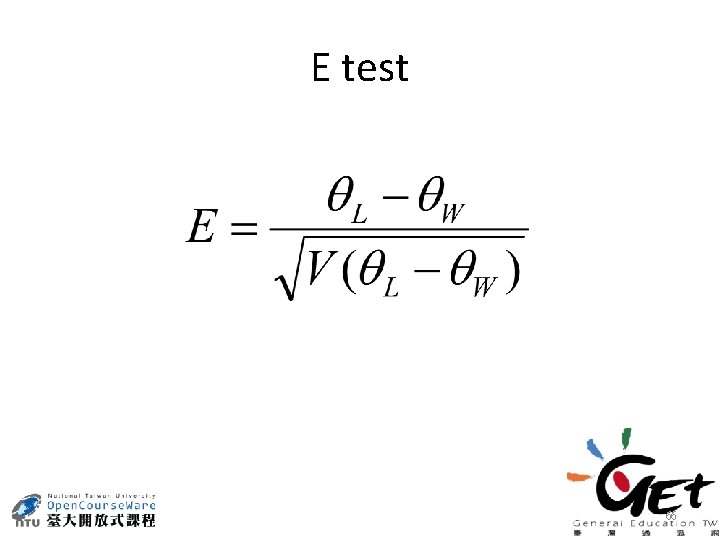
H test 65

E test 66

Positive selection (before fixation) During the process of frequency increase, population tends to have more high and low frequency variants, H and D are sensitive tests, but E is largely unaffected. Source: Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics 174: p. 1431. 67

Positive selection (after fixation) After fixation, the E-test quickly becomes the most powerful test, because high- frequency variations quickly go fixation. Source: Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics 174: p. 1431. 68

Population growth When a population increase in size, it tends to have an excess of low-frequency variants. Both D and E are sensitive to this type of deviation. E is the most sensitive test because highfrequency variants are the last to reach the new equilibrium after expansion. In contrast, H is unaffected. Source: Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics 174: p. 1431. 69

Population Shrinkage When a population decrease in size, the number of low-frequency variants tends to be smaller than expected. Thus, H can be sensitive to population shrinkage, whereas D and E are largely unaffected. Source: Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics 174: p. 1431. 70

Source: Rasmus Nielsen 2005. Molecular signatures of natural selection. Annual Review of Genetics 39: p. 197. 71

Copyright Declaration Work Licensing Author/Source Page P 3 National Taiwan University Chau-Ti Ting P 4 National Taiwan University Chau-Ti Ting “When the numbers of substitutions are small, the statistics is unlikely to follow the t distribution and we are likely to reject the null hypothesis. Another method was proposed” Dan Graur and Wen-Hsiung Li 2000. Fundamentals of Molecular Evolution. , p. 120. Sinauer Associates, Inc. Sunderland, MA, USA. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 15 Dan Graur and Wen-Hsiung Li 2000. Fundamentals of Molecular Evolution. , p. 120. Sinauer Associates, Inc. Sunderland, MA, USA. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 15 72

Work “Fig. 2. The number of nonsynonymous substitutions per … Similar results are obtained by using the method of Nei and Gojobori (53). (Swanson et al, 2001)” Licensing Author/Source Page Hurng-Yi Wang, Hua Tang, C. -K. James Shen, and Chung-I Wu 2003. Rapidly Evolving Genes in Human. I. The Glycophorins and Their Possible Role in Evading Malaria Parasites. . Molecular Biology and Evolution: 20: p. 1795. http: //mbe. oxfordjournals. org/content/20/11/1795. full. pdf+html 2012/0704 visited Note 1. P 16 Willie J. Swanson, Andrew G. Clark, Heidi M. Waldrip-Dail, Mariana F. Wolfner, and Charles F. Aquadro 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. . PNAS 98: p. 7375 http: //www. pnas. org/content/98/13/7375. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • PNAS Terms and Conditions for Use P 18 Note 1. OXFORD OPEN LICENCE AGREEMENT Authors who choose to participate in the Oxford Open initiative and pay to have their paper freely available online will be asked to sign an open access licence agreement which reflects the open access model outlined below. Articles published under the Oxford Open model are made freely available online immediately upon publication, as part of a long-term archive, without subscription barriers to access. We have chosen to implement the Creative Commons Attribution-Non Commercial licence for articles published under the Oxford Open model. 73

Work Licensing Author/Source Page P 20 Yoshiyuki Suzuki and Takashi Gojobori 1999. A Method for Detecting Positive Selection at Single Amino Acid Sites. Molecular Biology and Evolution 16: p. 1315. http: //mbe. oxfordjournals. org/content/16/10/1315. full. pdf+html 2012/0704 visited P 21 Yoshiyuki Suzuki and Takashi Gojobori 1999. A Method for Detecting Positive Selection at Single Amino Acid Sites. Molecular Biology and Evolution 16: p. 1315. http: //mbe. oxfordjournals. org/content/16/10/1315. full. pdf+html 2012/0704 visited Ziheng Yang 2012. PAML: Phylogenetic Analysis by Maximum Likelihood (User Guide), p. 29. http: //abacus. gene. ucl. ac. uk/software/paml. DOC. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 22 P 4 National Taiwan University Chau-Ti Ting 74

Work “Assuming ω follows discrete -gamma distribution, positive selection is tested using a likelihood ratio test comparing a null model that does not allow ω > 1 with an alternative model that does. “ Licensing Author/Source Ziheng Yang 2006. Computational Molecular Evolution, p. 274 -275. Oxford University Press. , London, UK. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. Ziheng Yang 2012. PAML: Phylogenetic Analysis by Maximum Likelihood (User Guide), p. 30. http: //abacus. gene. ucl. ac. uk/software/paml. DOC. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. “In branch test, positive selection is detected along the branch only if ω ratio average over all sites is significantly greater … The branch-site models attempt to detect signals of such local episodic natural selection. ” Ziheng Yang 2006. Computational Molecular Evolution, p. 279 -280. Oxford University Press. , London, UK. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. Ziheng Yang 2012. PAML: Phylogenetic Analysis by Maximum Likelihood (User Guide), p. 32. http: //abacus. gene. ucl. ac. uk/software/paml. DOC. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. Page P 24 P 25 P 26 P 27 75

Work Licensing Author/Source Page Andrew G. Clark, Stephen Glanowski, Rasmus Nielsen, Paul D. Thomas, Anish P 29 Kejariwal, Melissa A. Todd, David M. Tanenbaum, Daniel Civello, Fu Lu, Brian Murphy, Steve Ferriera, Gary Wang, Xianqgun Zheng, Thomas J. White, John J. Sninsky, Mark D. Adams, Michele Cargill 2003. Inferring Nonneutral Evolution from Human-Chimp-Mouse Orthologous Gene Trios. Science 302: p. 1960. http: //www. sciencemag. org/content/302/5652/1960. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. Andrew G. Clark, Stephen Glanowski, Rasmus Nielsen, Paul D. Thomas, Anish P 30 Kejariwal, Melissa A. Todd, David M. Tanenbaum, Daniel Civello, Fu Lu, Brian Murphy, Steve Ferriera, Gary Wang, Xianqgun Zheng, Thomas J. White, John J. Sninsky, Mark D. Adams, Michele Cargill 2003. Inferring Nonneutral Evolution from Human-Chimp-Mouse Orthologous Gene Trios. Science 302: p. 1960. http: //www. sciencemag. org/content/302/5652/1960. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Science AAAS Copyright Statement 76

Work Licensing Author/Source Margaret A. Bakewell, Peng Shi, and, Jianzhi Zhang 2007. More genes underwent positive selection in chimpanzee evolution than in human evolution. PNAS 104: p. 7489. http: //www. pnas. org/content/104/18/7489. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • PNAS Terms and Conditions for Use Dan Graur and Wen-Hsiung Li 2000. Fundamentals of Molecular Evolution. , p. 56. Sinauer Associates, Inc. Sunderland, MA, USA. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. Page P 34 P 35 P 36 P 39 77

Work Licensing Author/Source Page P 40 National Taiwan University Chau-Ti Ting John H. Mc. Donald and Martin Kreitman P 41 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: p. 652. http: //www. nature. com/nature/journal/v 351/n 6328/pdf/351652 a 0. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Nature Terms and Conditions Rasmus Nielsen P 43, P 44 2005. Molecular signatures of natural selection. Annual Review of Genetics 39: p. 197. http: //www. annualreviews. org/doi/full/10. 1146/annurev. genet. 39. 073003. 11242 0 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. “Heterozyosity (h): probability that two randomly chosen sequences from population are different … DNA sequences, it is more informative to consider the number of nucleotide differences between the two sequences. ” Wen-Hsiung Li 2006. Heterozygosity. Encyclopedia of Life Sciences. http: //onlinelibrary. wiley. com/doi/10. 1038/npg. els. 0005080/full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Wiley Online Library Terms and Conditions of Use • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 45 P 46 National Taiwan University Chau-Ti Ting 78

Work Licensing Author/Source Page P 47 National Taiwan University Chau-Ti Ting P 48 National Taiwan University Chau-Ti Ting “A segregating site (S) is a site that shows variation among the sequences in the sample. Under the infinitesite model, Watterson (1975) showed that the mean and variance are given by ” M. Prakash 2007. Molecular Genetics, p. 237. Discovery Publishing House, New Delhi, India. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 49 “Π, the average number of nucleotide differences … Πij is the number of nucleotide differences between the ith and jth sequences and n(n 1)/2 is the number of possible pairs” M. Prakash 2007. Molecular Genetics, p. 236. Discovery Publishing House, New Delhi, India. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 50 “The mutation model that is commonly used to study Π is the … Under this model and assumption of random mating, Watterson (1975) showed that the mean of Π is given by” M. Prakash 2007. Molecular Genetics, p. 236. Discovery Publishing House, New Delhi, India. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 51 79

Work Licensing Author/Source Page P 52 National Taiwan University Chau-Ti Ting P 53 National Taiwan University Chau-Ti Ting “K is strongly affected by the existence of deleterious mutations, … increase θπ considerably but have little effect on θw, i. e. D should have a positive sign. ” “Recent mutations are close to the tips (external branches) in the genealogy and therefore are mostly included … and θw are less affected than ηe by the presence of selection. “ M. Prakash 2007. Molecular Genetics, p. 246. Discovery Publishing House, New Delhi, India. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 55 Yun-Xin Fu and Wen-Hsiung Li 1993. Statistical Tests of Neutrality of Mutations. Genetics 133: p. 693. http: //www. genetics. org/content/133/3/693. full. pdf+html It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 56 M. Prakash 2007. Molecular Genetics, p. 246. Discovery Publishing House, New Delhi, India. It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 60 80

Work “During bottleneck, the number of low-frequency variants tends to be smaller than expected … in size, it tends to have an excess of low-frequency variants. ” Licensing Author/Source Page Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing highfrequency variants. Genetics 174: p. 1431. http: //www. genetics. org/content/174/3/1431. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 61 Justin C. Fay and Chung-I Wu 2000. Hitchhiking Under Positive Darwinian Selection. Genetics 155: p. 1405. http: //www. genetics. org/content/155/3/1405. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. Shiou-Hwei Yeh, Hurng-Yi Wang, Ching-Yi Tsai, Chuan-Liang Kao, Jyh. Yuan Yang, Hwan-Wun Liu, Ih-Jen Su, Shih-Feng Tsai, Ding-Shinn Chen, Pei. Jer Chen, and the National Taiwan University SARS Research Team. 2004. Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: Molecular epidemiology and genome evolution. PNAS 101: p. 2542. http: //www. pnas. org/content/101/8/2542. full. pdf+html It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • PNAS Terms and Conditions for Use P 63 P 64 81

Work Licensing Author/Source Page “During the process of frequency increase, population tends to have more high and low frequency variants, H and D are sensitive tests, but E is largely unaffected. ” Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing highfrequency variants. Genetics 174: p. 1431. http: //www. genetics. org/content/174/3/1431. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 67 “After fixation, the E-test quickly becomes the most powerful test, because high- frequency variations quickly go fixation. ” Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing highfrequency variants. Genetics 174: p. 1431. http: //www. genetics. org/content/174/3/1431. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 68 When a population increase in size, it tends to have an excess of low-frequency variants. … E is the most sensitive test because highfrequency variants are the last to reach the new equilibrium after expansion. In contrast, H is unaffected. Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing highfrequency variants. Genetics 174: p. 1431. http: //www. genetics. org/content/174/3/1431. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 69 “When a population decrease in size, the number of lowfrequency variants tends to be smaller than expected. Thus, H can be sensitive to population shrinkage, whereas D and E are largely unaffected. ” Kai Zeng, Yun-Xin Fu, Suhua Shi, and Chung-I Wu 2006. Statistical tests for detecting positive selection by utilizing highfrequency variants. Genetics 174: p. 1431. http: //www. genetics. org/content/174/3/1431. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. P 70 82

Work Licensing Author/Source Page Rasmus Nielsen P 71 2005. Molecular signatures of natural selection. Annual Review of Genetics 39: p. 197. http: //www. annualreviews. org/doi/full/10. 1146/annurev. genet. 39. 073003. 11242 0 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCWOct 29. pdf)" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act. 83
 Martin kreitman
Martin kreitman Similarities
Similarities Natural selection vs artificial selection
Natural selection vs artificial selection Difference between continuous and discontinuous variation
Difference between continuous and discontinuous variation Directional selection example
Directional selection example Natural selection vs artificial selection
Natural selection vs artificial selection Dsa vs rsa digital signature
Dsa vs rsa digital signature Beadgcf flats
Beadgcf flats Minimum distance classifier
Minimum distance classifier Compact multi-signatures for smaller blockchains
Compact multi-signatures for smaller blockchains Exchange 2007 signatures
Exchange 2007 signatures Uncitral model law on electronic signatures
Uncitral model law on electronic signatures Intruders use virus signatures fabricate
Intruders use virus signatures fabricate 15 key signatures
15 key signatures Uncitral model law on international commercial arbitration
Uncitral model law on international commercial arbitration Ocaml signatures
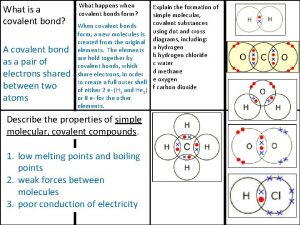
Ocaml signatures Melting and boiling point of oxygen
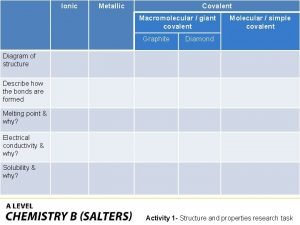
Melting and boiling point of oxygen Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Zinc oxide + nitric acid → zinc nitrate + water
Zinc oxide + nitric acid → zinc nitrate + water Chapter 15 section 1 darwins theory of natural selection
Chapter 15 section 1 darwins theory of natural selection Balancing selection vs stabilizing selection
Balancing selection vs stabilizing selection K selection r selection
K selection r selection K selection r selection
K selection r selection Two way selection and multiway selection in c
Two way selection and multiway selection in c Multiway selection in c
Multiway selection in c Mass selection and pure line selection
Mass selection and pure line selection P**n
P**n 4 principles of natural selection
4 principles of natural selection Stabilizing natural selection definition
Stabilizing natural selection definition Genetic drift
Genetic drift Overproduction in natural selection
Overproduction in natural selection Theory of natural selection
Theory of natural selection Aids basketball player
Aids basketball player 3 types of natural selection
3 types of natural selection Bird beak adaptation lab
Bird beak adaptation lab 3 types of natural selection
3 types of natural selection Natural selection definition biology
Natural selection definition biology Selección natural
Selección natural Natural selection
Natural selection Four steps to natural selection
Four steps to natural selection Macroevolution vs microevolution
Macroevolution vs microevolution Genetic drift vs gene flow vs natural selection
Genetic drift vs gene flow vs natural selection Natural selection examples
Natural selection examples 3 types of natural selection
3 types of natural selection Natural selection foldable
Natural selection foldable Natural selection vs evolution
Natural selection vs evolution The different types of natural selection
The different types of natural selection Artificial selection
Artificial selection 4 principles of natural selection
4 principles of natural selection Analougous structures
Analougous structures Evolution is gradual
Evolution is gradual Three types of selection
Three types of selection Criteria for natural selection
Criteria for natural selection Natural selection simulation at phet answers
Natural selection simulation at phet answers 3 types of natural selection
3 types of natural selection Basic principles of natural selection
Basic principles of natural selection Four steps to natural selection
Four steps to natural selection Natural selection menu
Natural selection menu Natural selection furniture
Natural selection furniture Natural selection requirements
Natural selection requirements Natural selection definition
Natural selection definition Basic principles of natural selection
Basic principles of natural selection Natural selection
Natural selection Overproduction in natural selection
Overproduction in natural selection Factors of natural selection
Factors of natural selection 4 principles of natural selection
4 principles of natural selection Picture of natural selection
Picture of natural selection Divergent evolution
Divergent evolution Selection pressure
Selection pressure What is stabilizing selection
What is stabilizing selection Stabilizing selection example
Stabilizing selection example Natural selection examples
Natural selection examples Natural selection
Natural selection Natural selection
Natural selection Modification
Modification Natural selection
Natural selection Natural selection examples
Natural selection examples Criteria for natural selection
Criteria for natural selection Evolution graphic organizer
Evolution graphic organizer Steiner tunnel test classifications
Steiner tunnel test classifications Evolution jeopardy review game
Evolution jeopardy review game How do geological processes affect evolution
How do geological processes affect evolution Resource partitioning tends to lead to a high degree of
Resource partitioning tends to lead to a high degree of Natural selection picture
Natural selection picture