Chapter 5 Genetic Linkage and Chromosome Mapping Jones






- Slides: 18

Chapter 5 Genetic Linkage and Chromosome Mapping Jones and Bartlett Publishers © 2005

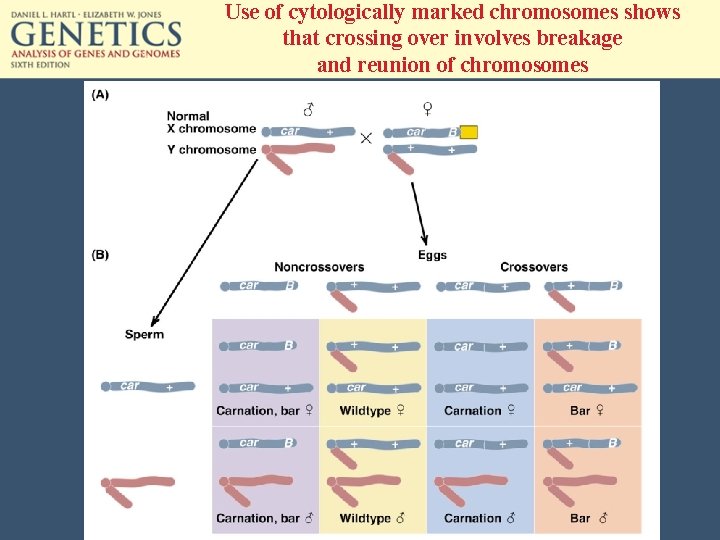
Use of cytologically marked chromosomes shows that crossing over involves breakage and reunion of chromosomes

Recombination rates in males and females • In Drosophila, males do not have any recombination, so all syntenic genes (those on one chromosome) always are completely linked. • How and why are not known. • Human males have recombination rates about 60% of that seen in females.

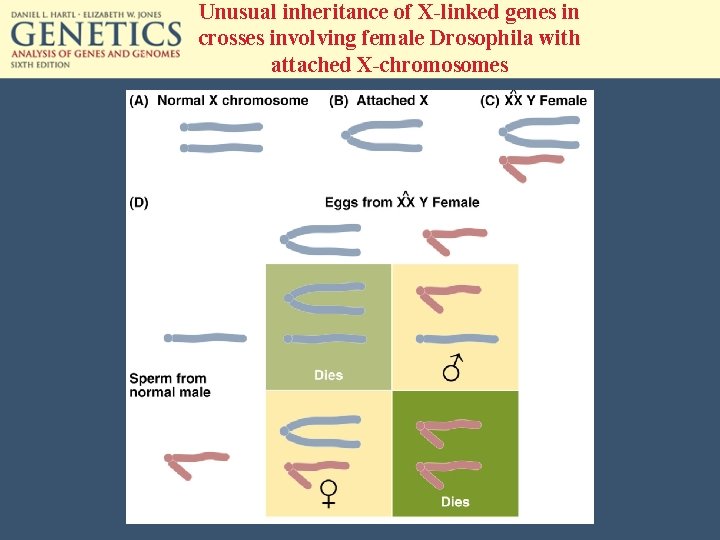
Unusual inheritance of X-linked genes in crosses involving female Drosophila with attached X-chromosomes

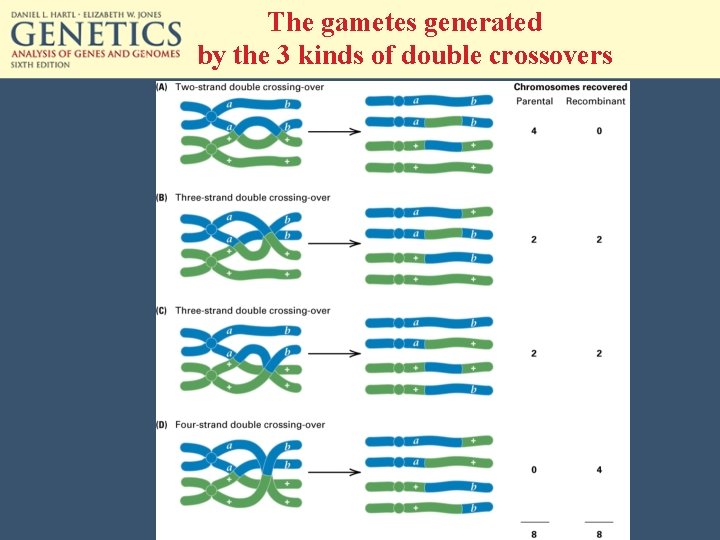
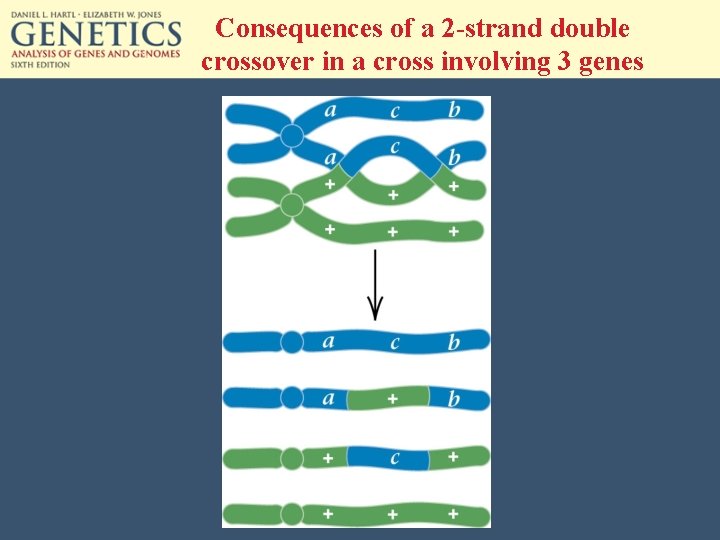
The gametes generated by the 3 kinds of double crossovers

Consequences of a 2 -strand double crossover in a cross involving 3 genes

Chromatid interference • Sometimes crossing over at one point on a chromosome interferes with other crossing over events on the same chromosome: • Chromatid interference means that there will be fewer double, triple, etc. crossing over events. • Interference is greatest over short distances.

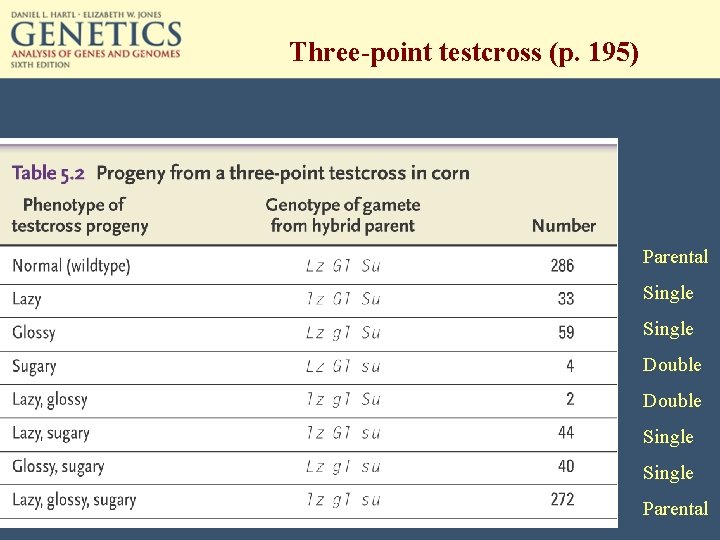
Three-point testcross (p. 195) Parental Single Double Single Parental

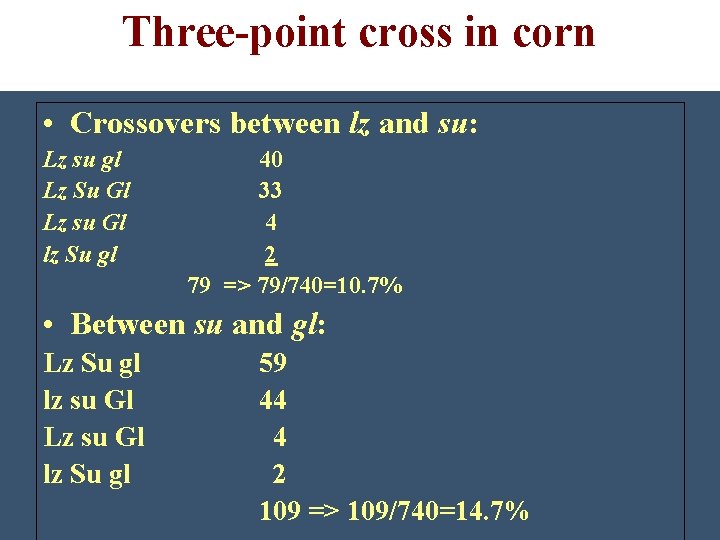
Three-point cross in corn • Crossovers between lz and su: Lz su gl Lz Su Gl Lz su Gl lz Su gl 40 33 4 2 79 => 79/740=10. 7% • Between su and gl: Lz Su gl lz su Gl Lz su Gl lz Su gl 59 44 4 2 109 => 109/740=14. 7%

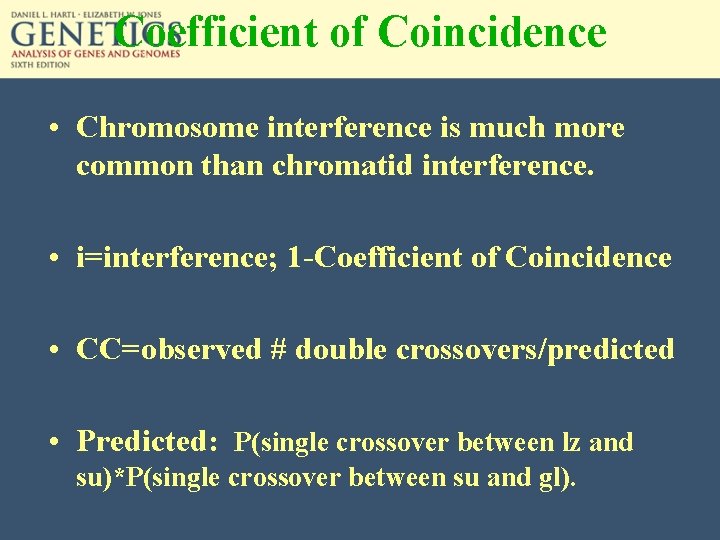
Coefficient of Coincidence • Chromosome interference is much more common than chromatid interference. • i=interference; 1 -Coefficient of Coincidence • CC=observed # double crossovers/predicted • Predicted: P(single crossover between lz and su)*P(single crossover between su and gl).

Coefficient of Coincidence For the previous corn data, R 1=0. 107 for lz and su R 2=0. 147 for su and gl. If independent, double crossovers would occur (R 1 x R 2)x # of progeny: 0. 107 x 0. 147 X 740=11. 6. Only 6 double crossovers were observed. CC=6/11. 6=0. 51, i=interference=1 -0. 51=0. 49.

Coefficient of Coincidence • With greater distance between genes, interference usually disappears. • In Drosophila, i=0 at about 10 c. M • For most organisms, interference disappears at about 30 c. M (CC=1).

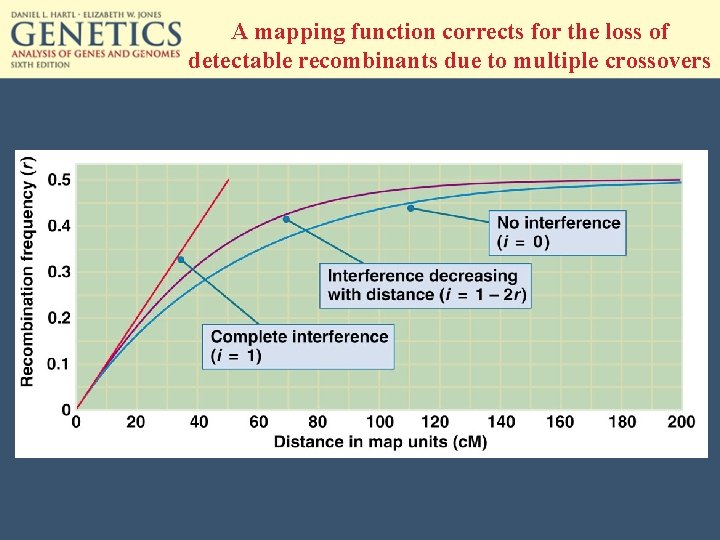
A mapping function corrects for the loss of detectable recombinants due to multiple crossovers

Three-Point Testcross Another example (Morgan’s data) Progeny Phenotype Genotype Scute echinus crossveinless sc ec cv /sc ec cv 4 Wild type + + + / sc ec cv 1 Scute sc + + / sc ec cv 997 Echinus crossveinless + ec cv /sc ec cv 1002 Scute echinus sc ec + / sc ec cv 681 Crossveinless + + cv / sc ec cv 716 Scute crossveinless sc + cv / sc ec cv 8808 Echinus + ec + / sc ec cv 8576 Total Number 20, 785

Morgan’s three-point results • Distance between sc and ec: (681 + 716 + 4 + 1)/20, 785 x 100 = 6. 74 c. M Distance between cv and ec: (1002 + 997 + 4 + 1)/20, 785 x 100 = 9. 65 c. M Predicted DC=(0. 0674 x 0. 0965) x 20, 785=135. 2 CC=5/135. 2 = 0. 0369; i = 0. 9630

Another example: • • Three linked loci in tomato: Mottled (m) vs. normal (M) leaf Smooth (P) vs. pubescent (p) epidermis Purple (Aw) vs. green (aw) stems

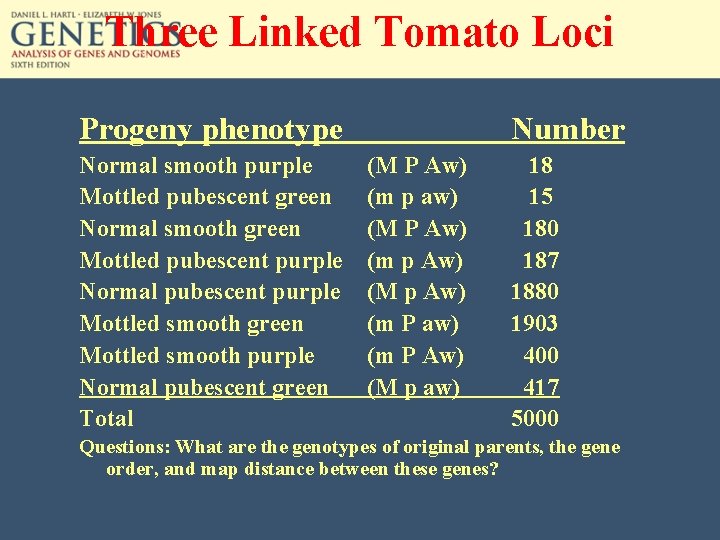
Three Linked Tomato Loci Progeny phenotype Normal smooth purple Mottled pubescent green Normal smooth green Mottled pubescent purple Normal pubescent purple Mottled smooth green Mottled smooth purple Normal pubescent green Total Number (M P Aw) (m p aw) (M P Aw) (m p Aw) (M p Aw) (m P aw) (m P Aw) (M p aw) 18 15 180 187 1880 1903 400 417 5000 Questions: What are the genotypes of original parents, the gene order, and map distance between these genes?

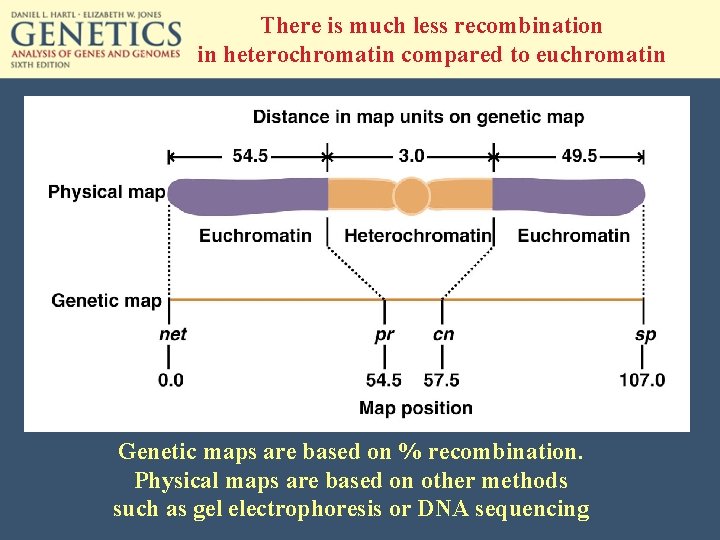
There is much less recombination in heterochromatin compared to euchromatin Genetic maps are based on % recombination. Physical maps are based on other methods such as gel electrophoresis or DNA sequencing