Chapter 3 Enzymes Dept of biochemistry and molecular

Chapter 3 Enzymes Dept. of biochemistry and molecular biology Hui-Qing Yuan
Key terms*** Ø Active site of enzymes (酶的活性中心) Ø Allosteric enzymes and allosteric regulation of enzymes (变构酶与酶的变构调节) Ø Covalent modification of enzymes (酶的共价修饰) Ø Isoenzymes (同 酶) Ø Zymogens and activation of zymogens (酶原与酶原激活)
Outline and key points Enzyme structure* and the active site*** How an enzyme facilitates the reaction? *** Quantitative analysis of E-catalyzed reaction rate *** Enzyme activity can be regulated***
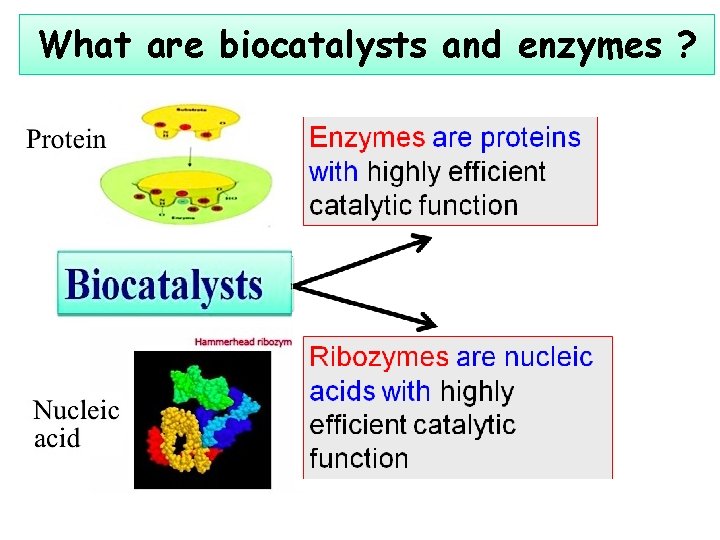
What are biocatalysts and enzymes ?
Biocatalysts and enzymes Ø Biocatalysts: catalyze every biological reactions --- enzymes --- ribozymes(核酶) and deoxyribozymes (脱氧核酶) Ø Enzymes typically are very large proteins, having highly efficient catalytic function Ø Ribozyme and deoxyribozyme: RNA and DNA with catalytic function
Section I Molecular structure and function of enzymes Ø Compositions of enzymes Ø Cofactors and coenzymes—辅助因子和辅酶*** Ø Active sites of enzymes---酶的活性中心*** Ø Isozymes---同 酶***
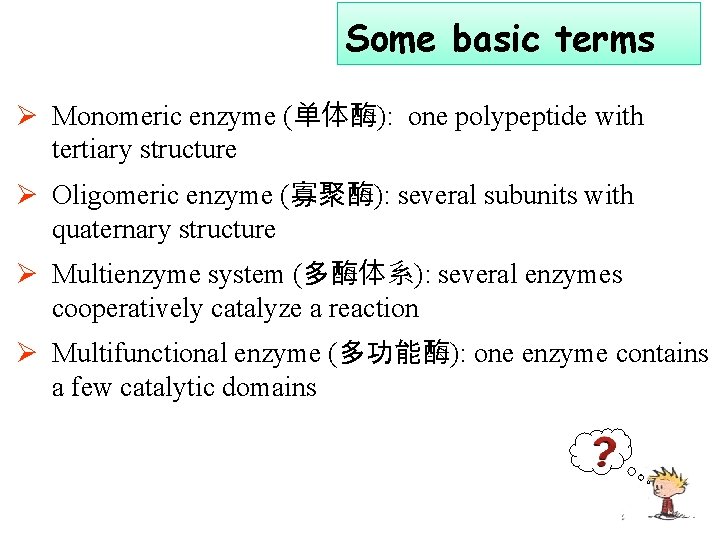
Some basic terms Ø Monomeric enzyme (单体酶): one polypeptide with tertiary structure Ø Oligomeric enzyme (寡聚酶): several subunits with quaternary structure Ø Multienzyme system (多酶体系): several enzymes cooperatively catalyze a reaction Ø Multifunctional enzyme (多功能酶): one enzyme contains a few catalytic domains
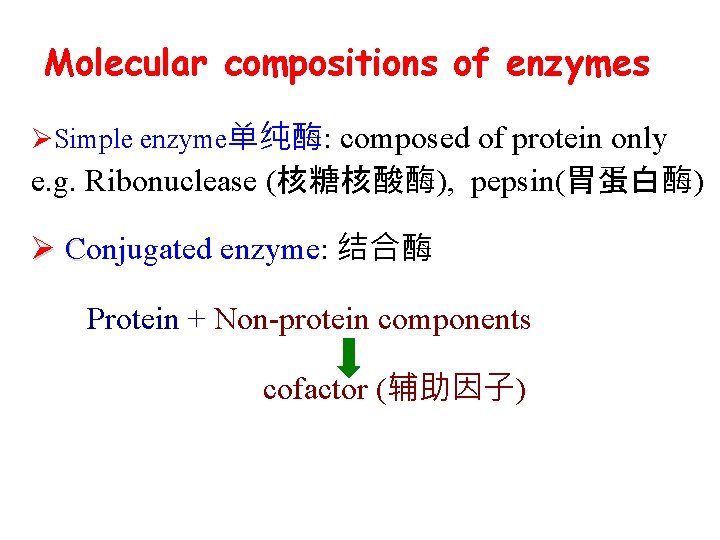
Molecular compositions of enzymes ØSimple enzyme单纯酶: composed of protein only e. g. Ribonuclease (核糖核酸酶), pepsin(胃蛋白酶) Ø Conjugated enzyme: 结合酶 Protein + Non-protein components cofactor (辅助因子)
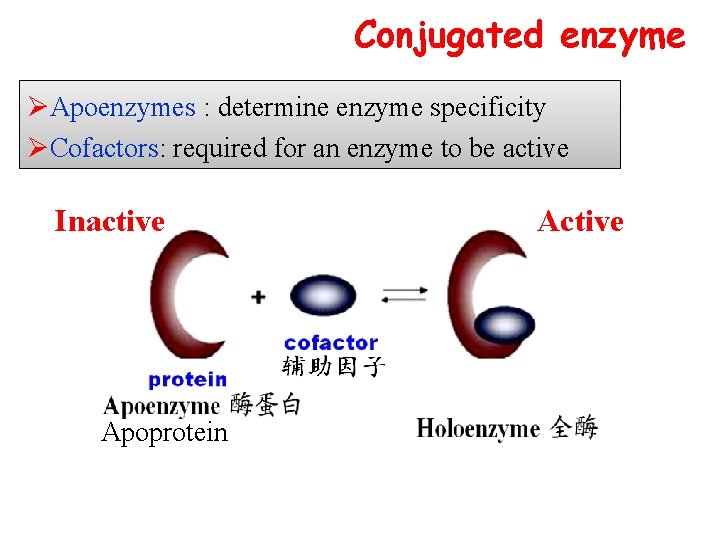
Conjugated enzyme ØApoenzymes : determine enzyme specificity ØCofactors: required for an enzyme to be active Inactive Apoprotein Active

Cofactors Ø As a constituent of enzyme 酶的组成成分 Ø Function: activation of an enzyme Ø Classified as: -- coenzymes (辅酶) -- prosthetic groups (辅基)
Cofactors Ø Coenzymes --loosely bind to apoproteins --act as transient carriers of specific functional groups Ø Prosthetic groups -- tightly or even covalently bound to the apoproteins -- essential for a complete, catalytically active enzyme What are cofactors? metal ions or small organic molecules
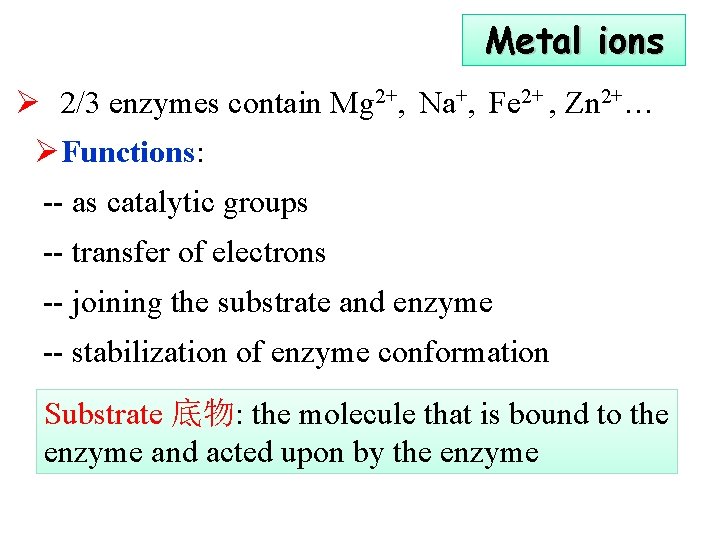
Metal ions Ø 2/3 enzymes contain Mg 2+, Na+, Fe 2+ , Zn 2+… ØFunctions: -- as catalytic groups -- transfer of electrons -- joining the substrate and enzyme -- stabilization of enzyme conformation Substrate 底物: the molecule that is bound to the enzyme and acted upon by the enzyme

ØMetalloenzyme (金属酶): ions bind tightly to E ØMetal activated enzyme (金属激活酶):ions loosely bind to apoenzyme, easy to be removed
Small organic molecules Ø Functions: transport of electrons, H+ and chemical groups (-COOH, -CH 3, -NH 2…) Ø Common components: water-soluble vitamins and nucleotides
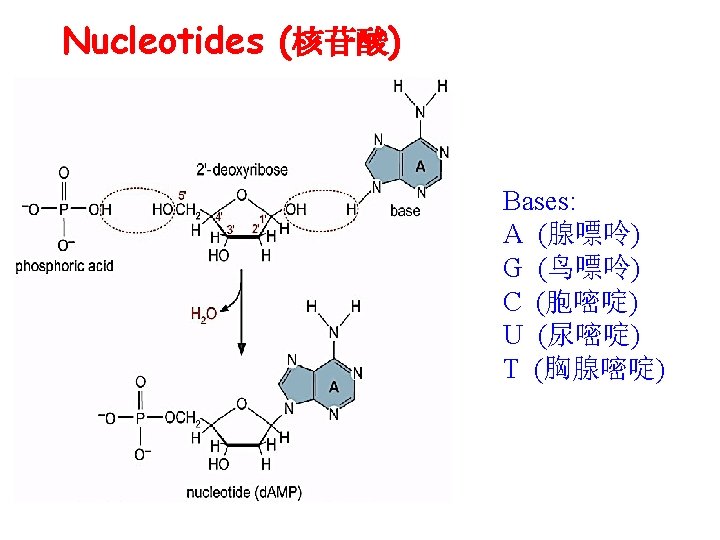
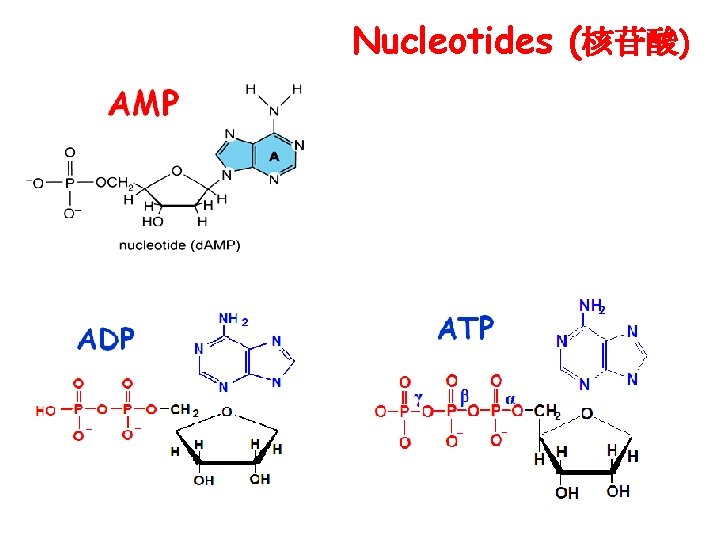
Nucleotides (核苷酸)

Vitamins Ø Essential nutrients Ø lipid- soluble vitamins: A, D, E , K Ø water- soluble vitamins are main components of cofactors B 1, B 2, B 6, B 12, C, Vitpp, biotin(生物素), pantothenic acid(泛酸), folic acid (叶酸) Chapter 5
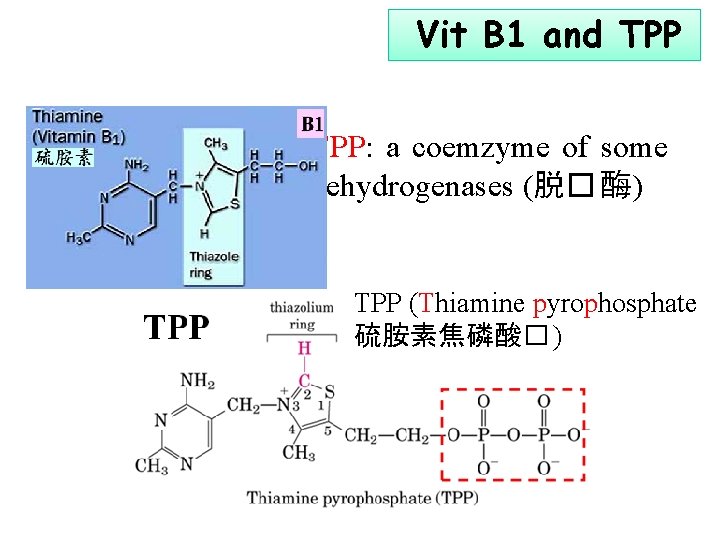
Vit B 1 and TPP: a coemzyme of some dehydrogenases (脱� 酶) TPP (Thiamine pyrophosphate 硫胺素焦磷酸� )
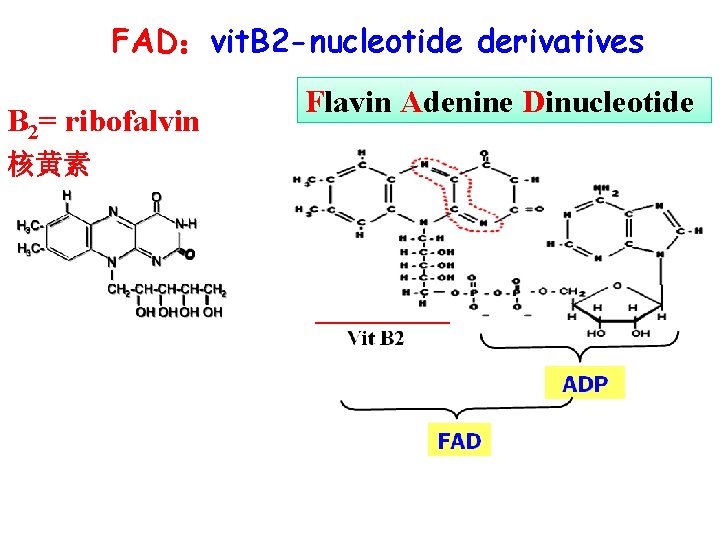
FAD:vit. B 2 -nucleotide derivatives B 2= ribofalvin 核黄素 Flavin Adenine Dinucleotide
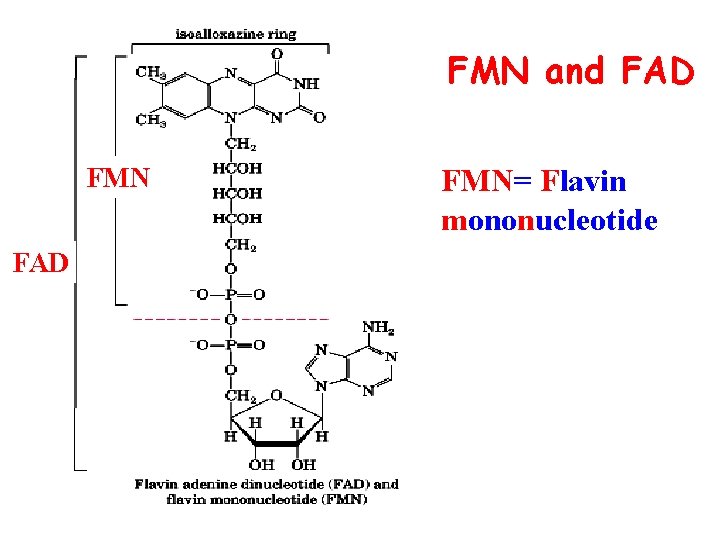
FMN and FAD FMN= Flavin mononucleotide
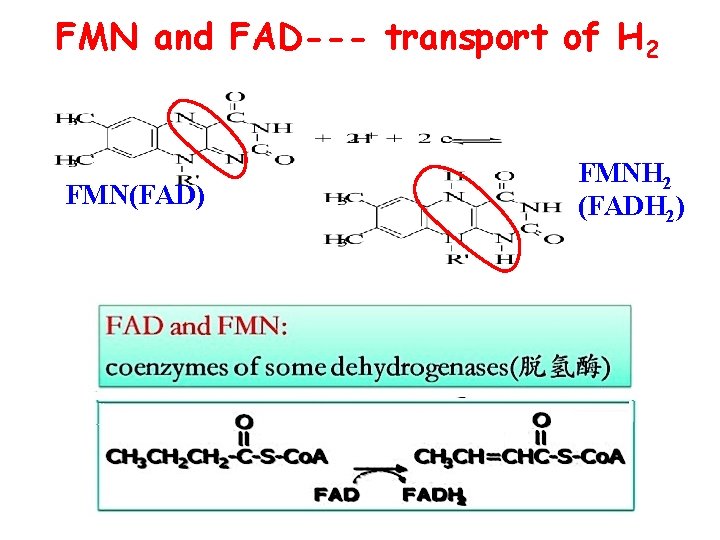
FMN and FAD--- transport of H 2 FMN(FAD) FMNH 2 (FADH 2)
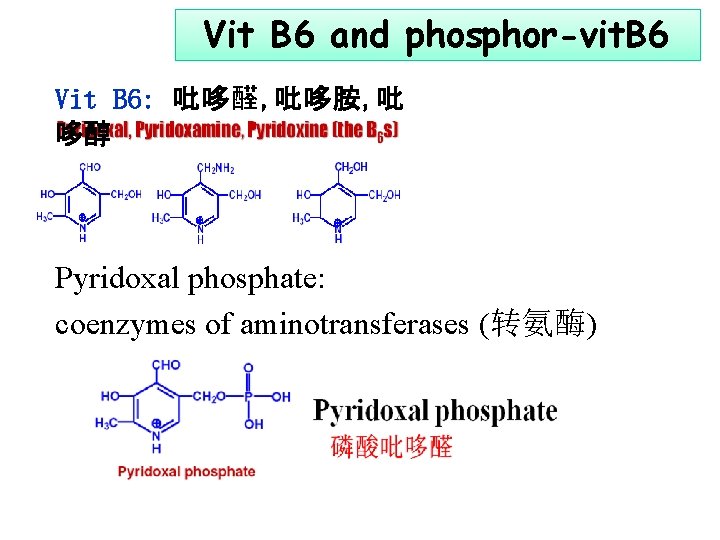
Vit B 6 and phosphor-vit. B 6 Vit B 6: 吡哆醛, 吡哆胺, 吡 哆醇 Pyridoxal phosphate: coenzymes of aminotransferases (转氨酶)
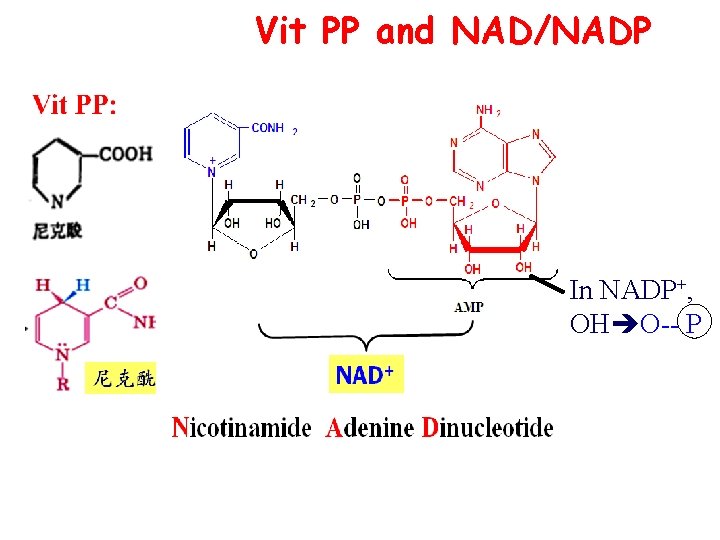
Vit PP and NAD/NADP In NADP+, OH O-- P
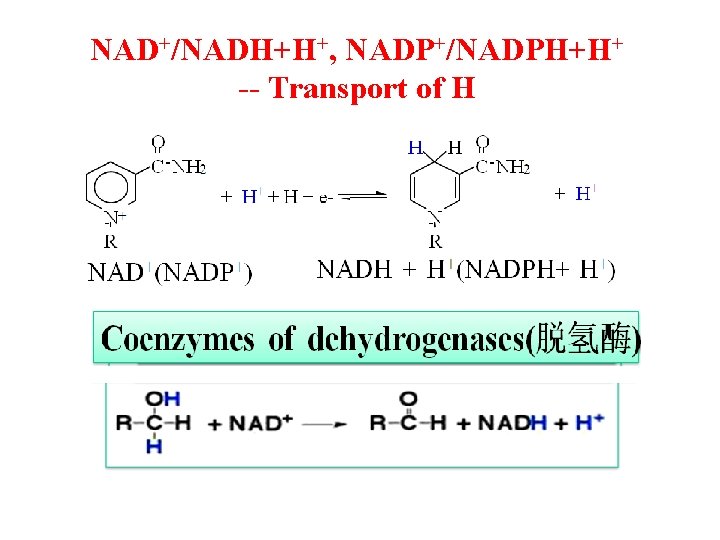
NAD+/NADH+H+, NADP+/NADPH+H+ -- Transport of H
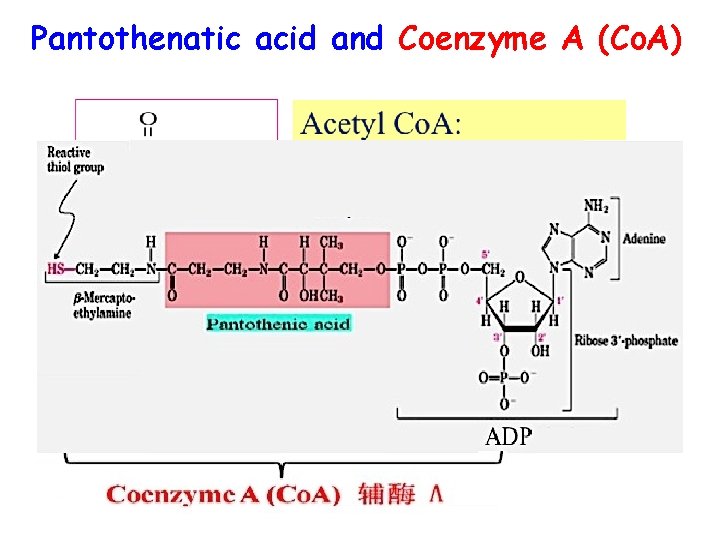
Pantothenatic acid and Coenzyme A (Co. A)
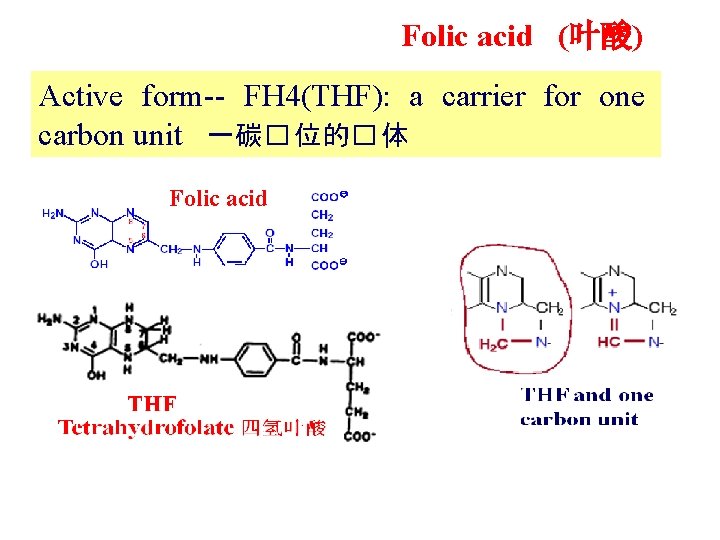
Folic acid (叶酸) Active form-- FH 4(THF): a carrier for one carbon unit 一碳� 位的� 体 Folic acid

Summary ØCofactors: --required for an E (structure and activity) --coenzymes, prosthetic groups -- metal ions: Zn 2+, Fe 2+…… --organic molecules: TPP, FMN/FMNH 2, FAD/FADH 2, NAD/NADH……
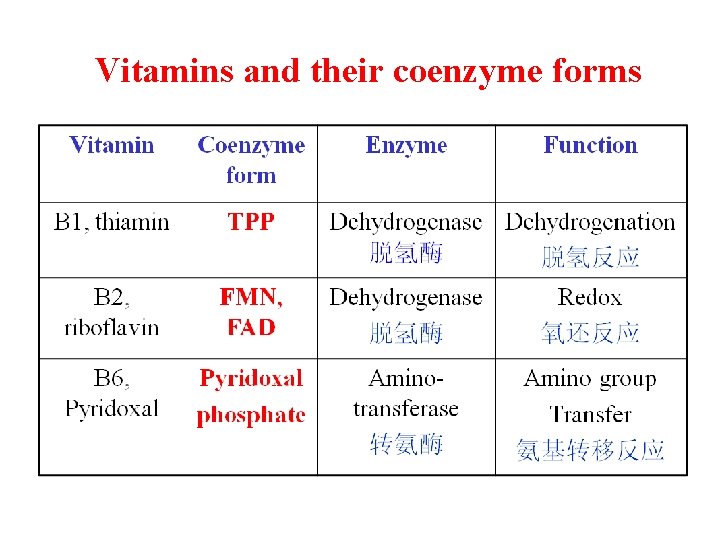
Vitamins and their coenzyme forms
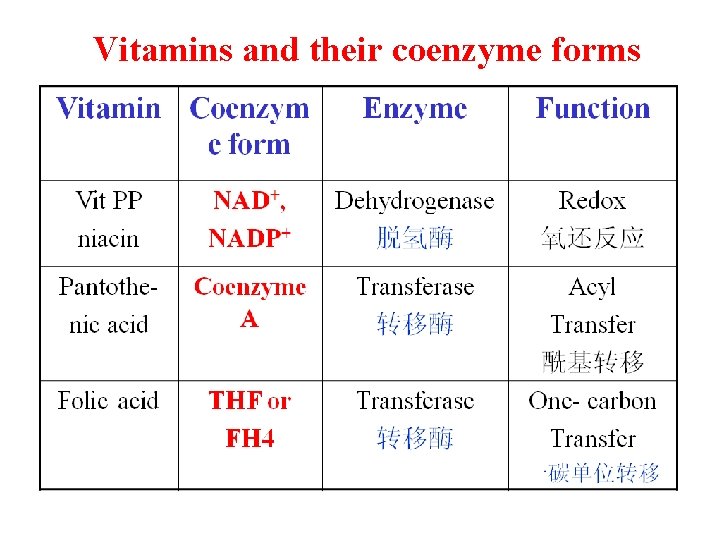
Vitamins and their coenzyme forms
How enzymes act as catalysts? Enzyme(E)-catalyzed reactions are characterized by: Ø Binding to the substrates (S) Ø conversion of S to a product How an enzyme binds to its substrate(s) ?
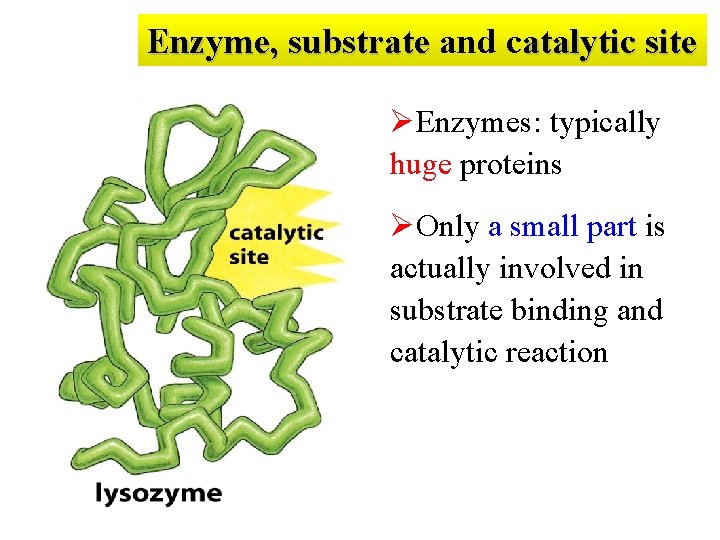
Enzyme, substrate and catalytic site ØEnzymes: typically huge proteins ØOnly a small part is actually involved in substrate binding and catalytic reaction
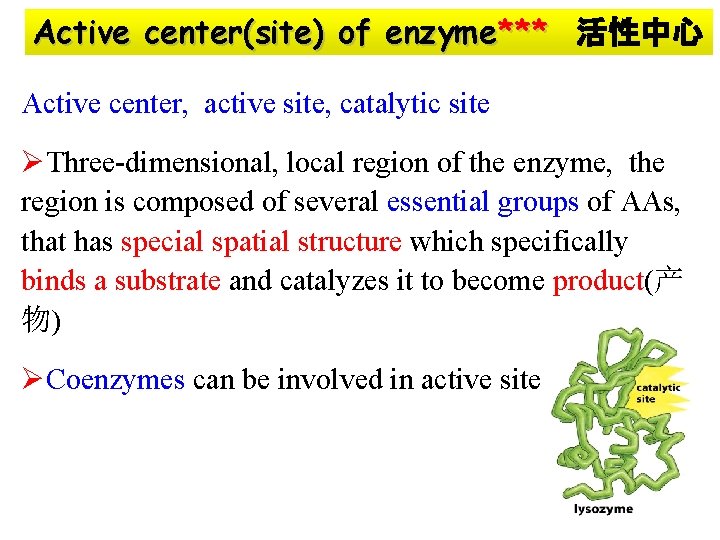
Active center(site) of enzyme*** 活性中心 Active center, active site, catalytic site ØThree-dimensional, local region of the enzyme, the region is composed of several essential groups of AAs, that has special spatial structure which specifically binds a substrate and catalyzes it to become product(产 物) ØCoenzymes can be involved in active site
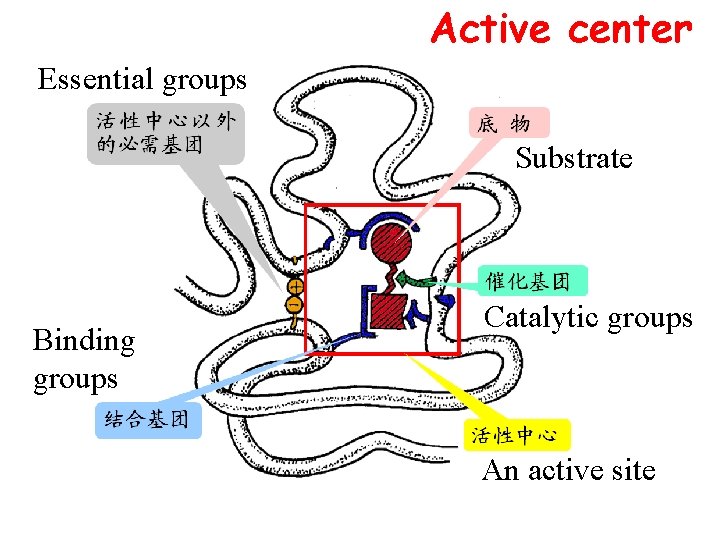
Active center Essential groups Substrate Binding groups Catalytic groups An active site

Essential groups*** (必需基� ) Ø Essential groups in the active sites --binding groups: bind S and coenzyme, and form an ES complex -- catalytic groups: catalyze the S to become product -- -OH, -COOH, -SH are usually essential groups in the active site ØEssential groups outside of the active sites maintenance of the spatial structure of active sites
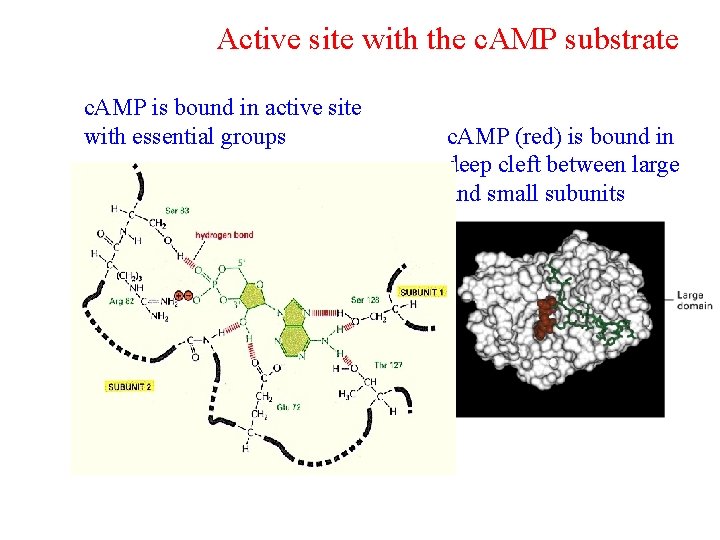
Active site with the c. AMP substrate c. AMP is bound in active site with essential groups c. AMP (red) is bound in deep cleft between large and small subunits
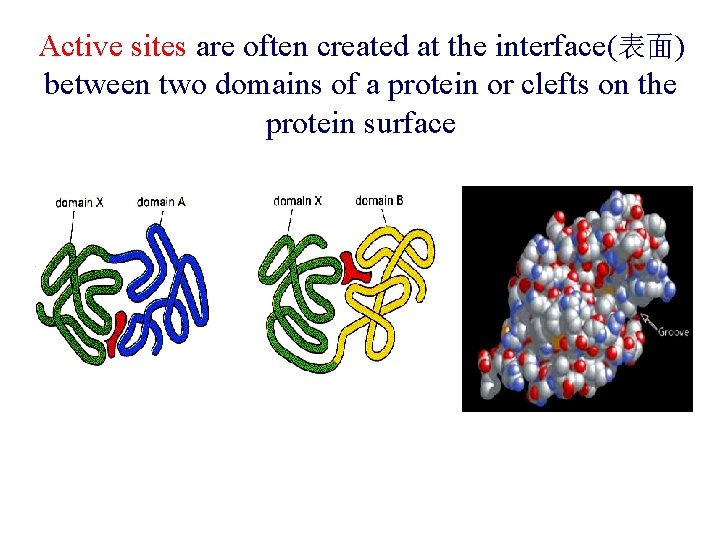
Active sites are often created at the interface(表面) between two domains of a protein or clefts on the protein surface
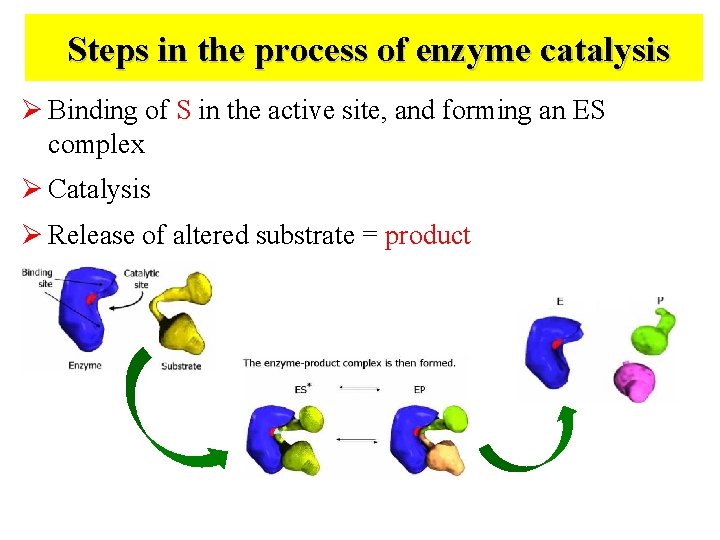
Steps in the process of enzyme catalysis Ø Binding of S in the active site, and forming an ES complex Ø Catalysis Ø Release of altered substrate = product
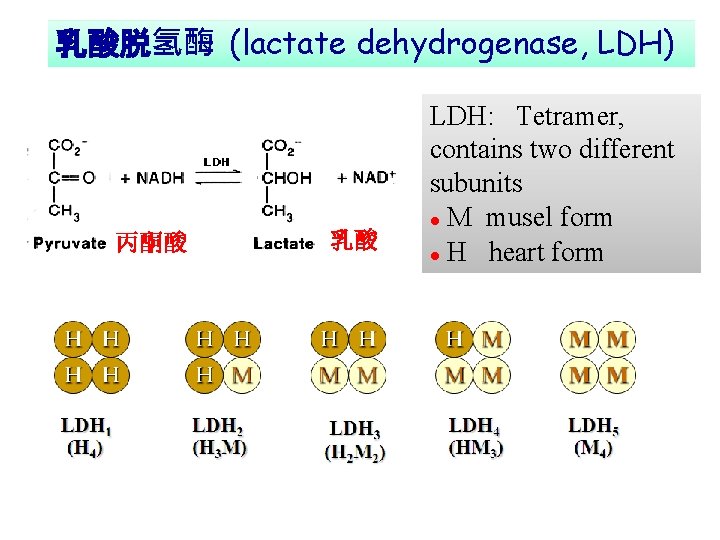
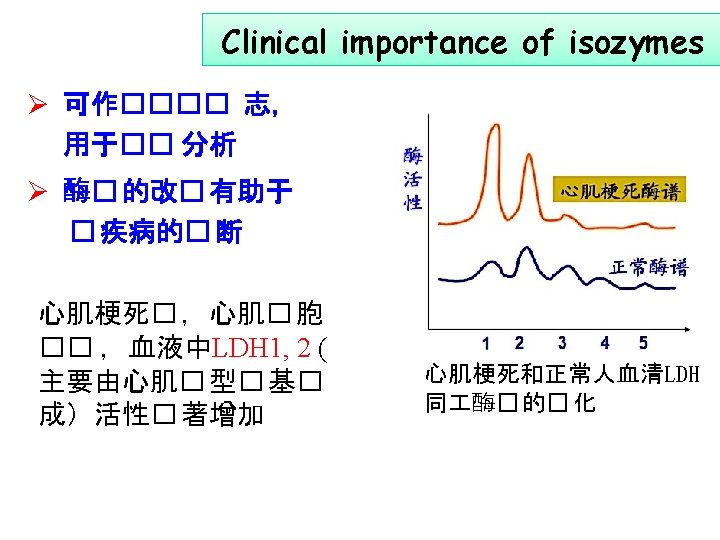
乳酸脱氢酶 (lactate dehydrogenase, LDH) 丙酮酸 乳酸 LDH: Tetramer, contains two different subunits l M musel form l H heart form
Isozymes (isoenzymes, 同 酶) A group of enzymes: Ø catalyze the same reaction with similar structure of active site Ø have different protein structure, physicochemical and immunological properties Ø Encoded by different genes or different transcripts from the same gene Ø vary in different tissues
Summary Ø In addition to catalytic RNAs/DNAs, all known enzymes are proteins Ø Many enzymes require cofactors for their catalysis Ø Water-soluble vitamins are main components of coenzymes or prosthetic groups Ø Substrate binding occurs in a pocket on the enzyme called the active site Ø E-catalyzed reactions are characterized by the formation of an ES complex

Section Ⅱ Mechanisms of Enzyme Action How an enzyme works? It increases the reaction rate by lowing activation energy

Enzymes, like other catalysts (催化剂): Ø increase the rate of reactions without themselves being altered in the process Ø do not determine the direction of a reaction Ø cannot alter the position of the equilibrium point 不改变反应的平衡常数
Characteristics of enzyme-catalyzed reaction* Enzymes, unlike other catalysts: Ø High efficiency (高效) Ø High specificity (特异性强) Ø Enzyme activity can be regulated(活性可�� ) Ø Enzyme activity is lost once the enzyme is denatured
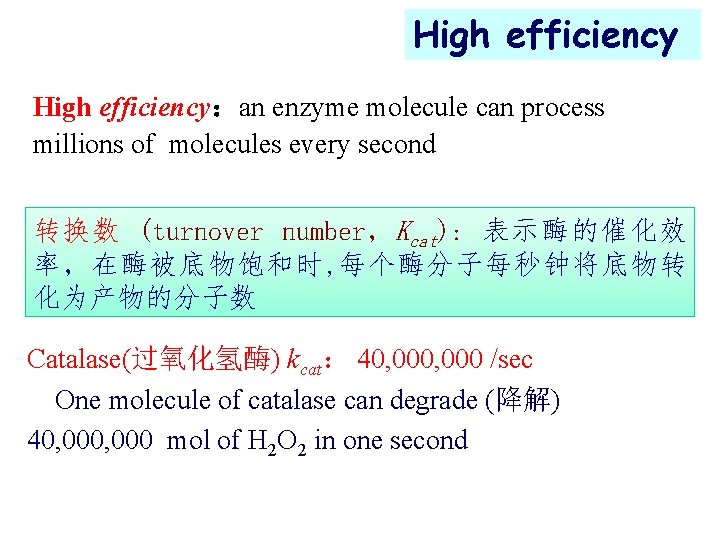
High efficiency:an enzyme molecule can process millions of molecules every second 转换数 (turnover number, Kcat): 表示酶的催化效 率,在酶被底物饱和时, 每个酶分子每秒钟将底物转 化为产物的分子数 Catalase(过氧化氢酶) kcat: 40, 000 /sec One molecule of catalase can degrade (降解) 40, 000 mol of H 2 O 2 in one second
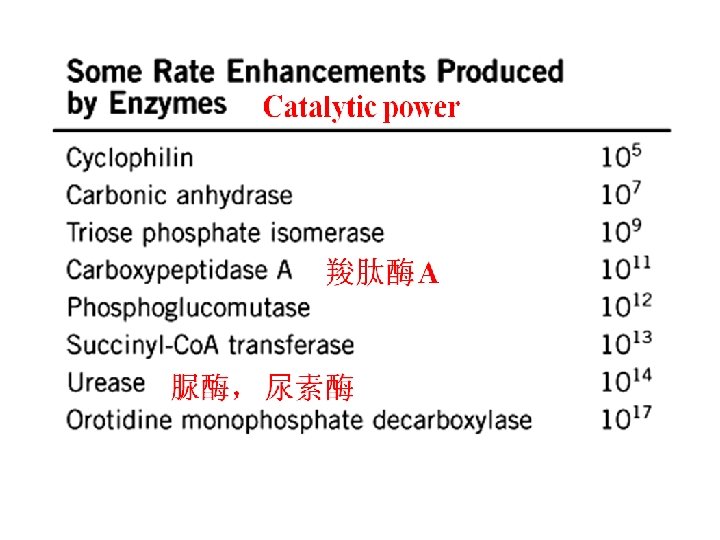
High specificity Ø The selectivity(选择性) of an enzyme for the substrate molecule which is catalyzed Ø Specificity depends on mutual(双方) structural recognition between enzyme (E) and substrate(S)
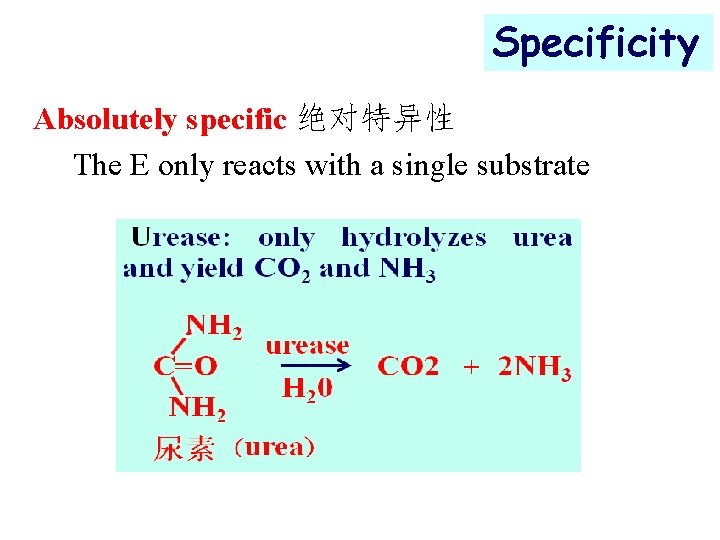
Specificity Absolutely specific 绝对特异性 The E only reacts with a single substrate
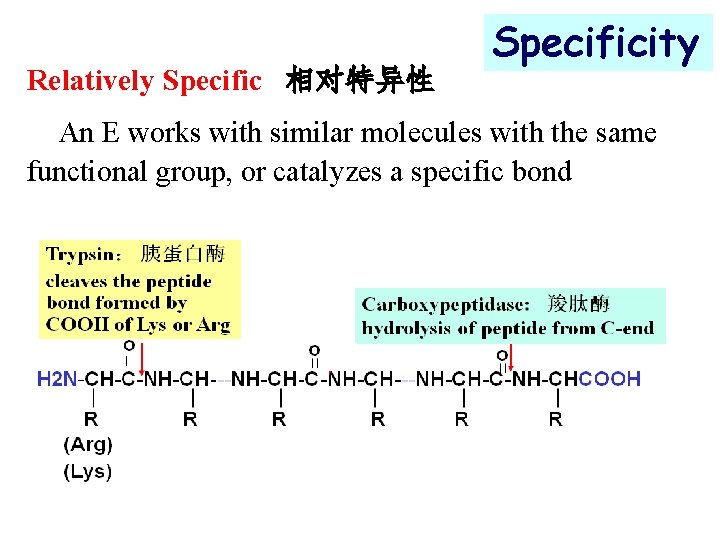
Relatively Specific 相对特异性 Specificity An E works with similar molecules with the same functional group, or catalyzes a specific bond

Specificity Stereochemically specific 立体结构特异性 An E works with the proper stereo-isomer of a substrate
Regulation of enzyme activity Enzyme activity can be regulated by: Ø Altering the enzyme structure, or quantity Ø Changing the interaction of S to the E Ø Changing the product quantity Ø Others: H+, temperature…
After binding to a substrate, why the enzyme is able to accelerate the chemical reaction? Energy barrier 能障
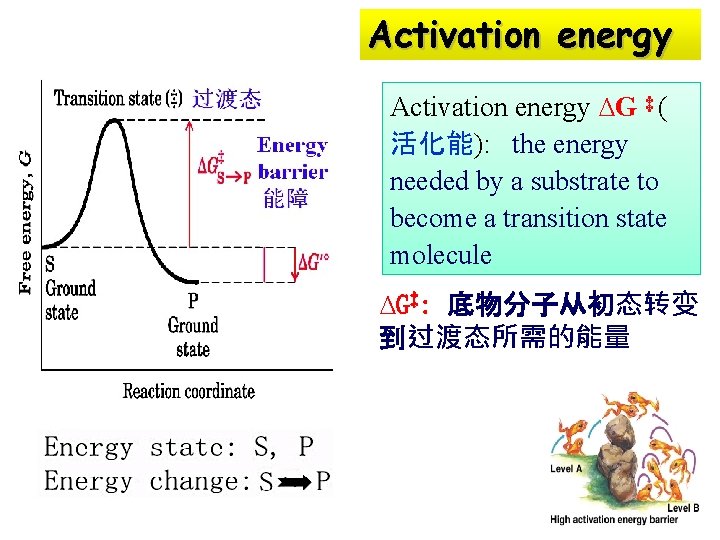
Activation energy ∆G ‡ ( 活化能): the energy needed by a substrate to become a transition state molecule ∆G‡: 底物分子从初态转变 到过渡态所需的能量
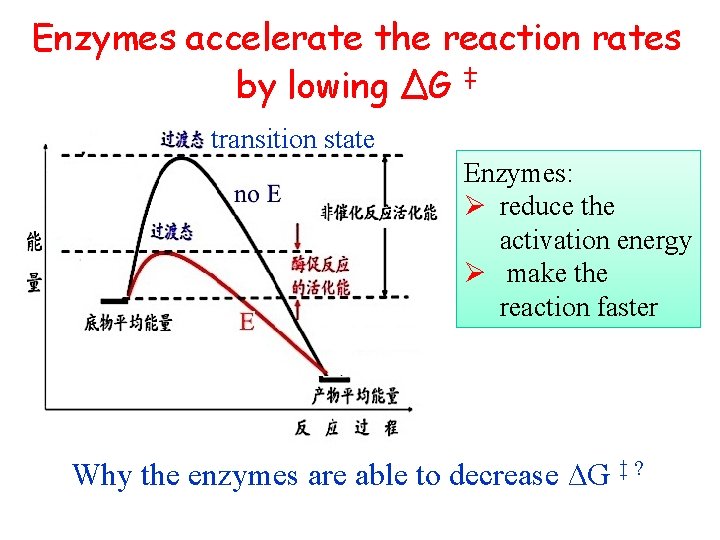
Enzymes accelerate the reaction rates by lowing ∆G ‡ transition state Enzymes: Ø reduce the activation energy Ø make the reaction faster Why the enzymes are able to decrease ∆G ‡ ?
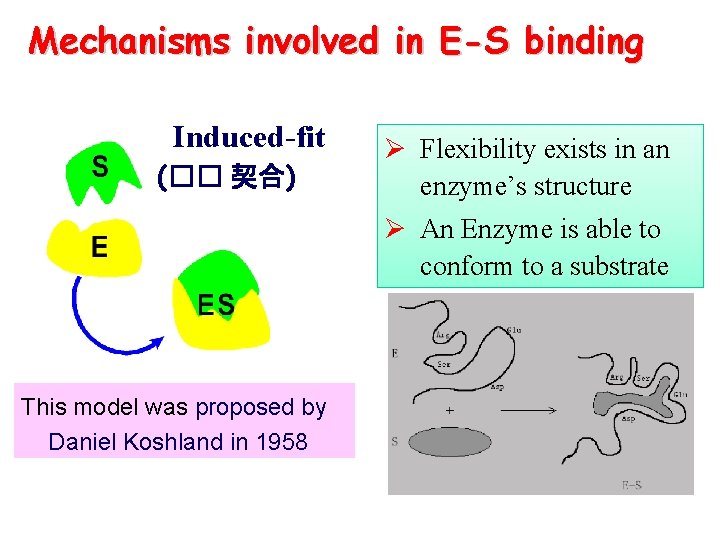
Mechanisms involved in E-S binding Induced-fit (�� 契合) Ø Flexibility exists in an enzyme’s structure Ø An Enzyme is able to conform to a substrate This model was proposed by Daniel Koshland in 1958
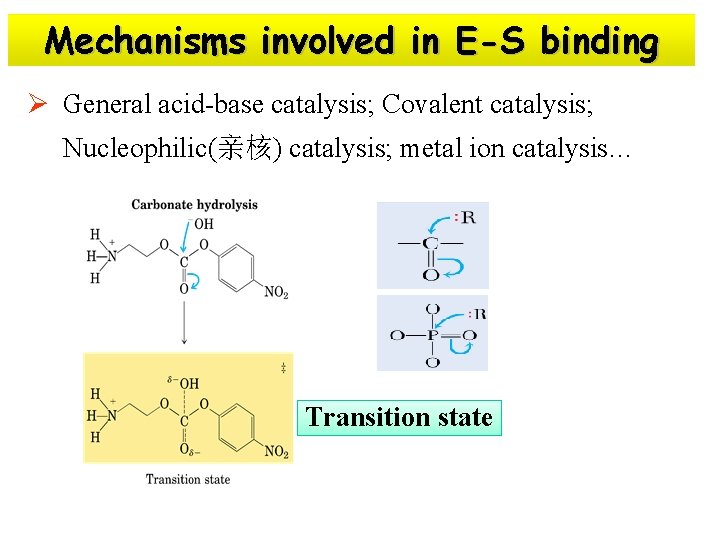
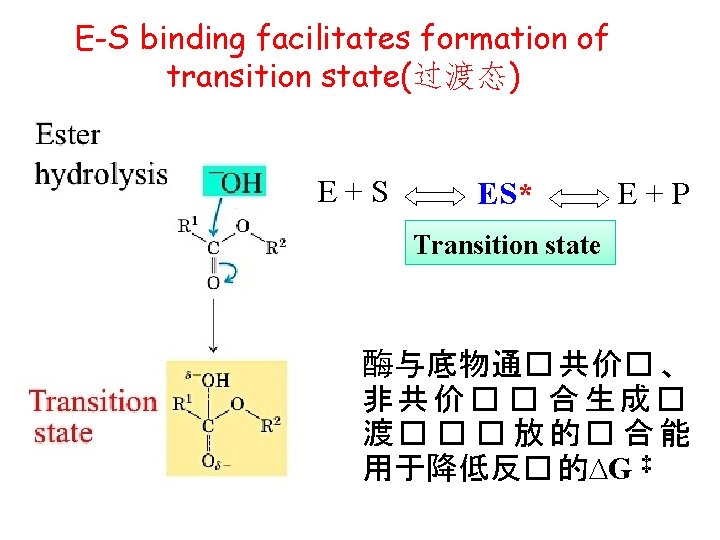
Mechanisms involved in E-S binding Ø General acid-base catalysis; Covalent catalysis; Nucleophilic(亲核) catalysis; metal ion catalysis… Transition state
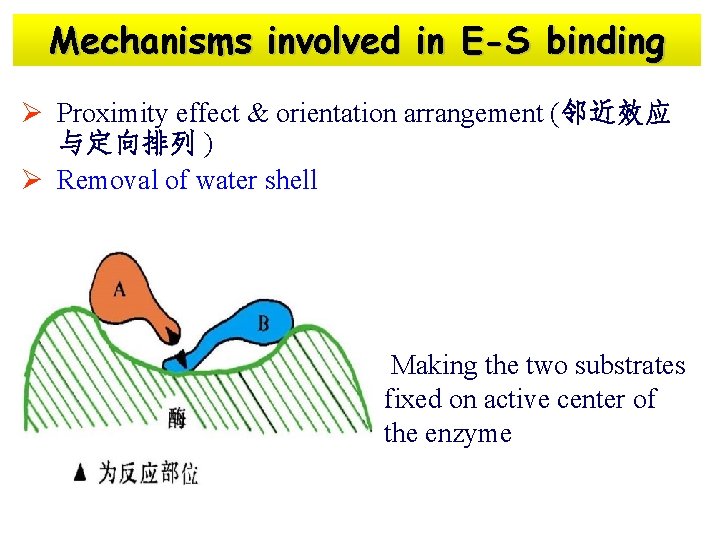
Mechanisms involved in E-S binding Ø Proximity effect & orientation arrangement (邻近效应 与定向排列 ) Ø Removal of water shell Making the two substrates fixed on active center of the enzyme
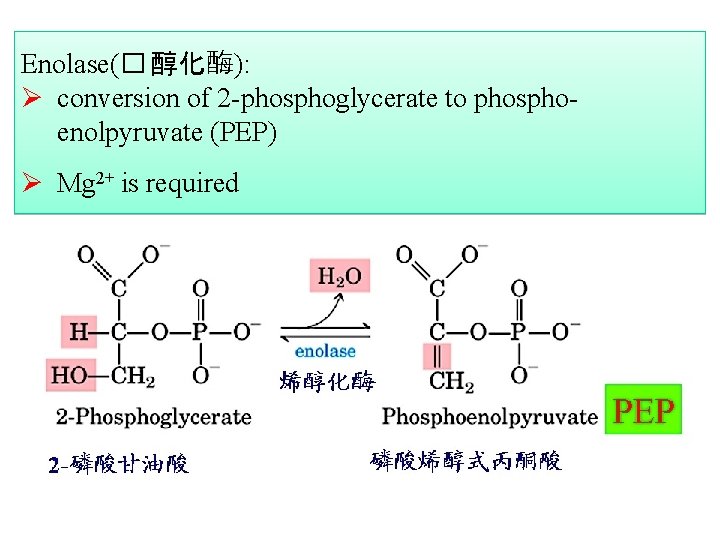
Enolase(� 醇化酶): Ø conversion of 2 -phosphoglycerate to phosphoenolpyruvate (PEP) Ø Mg 2+ is required
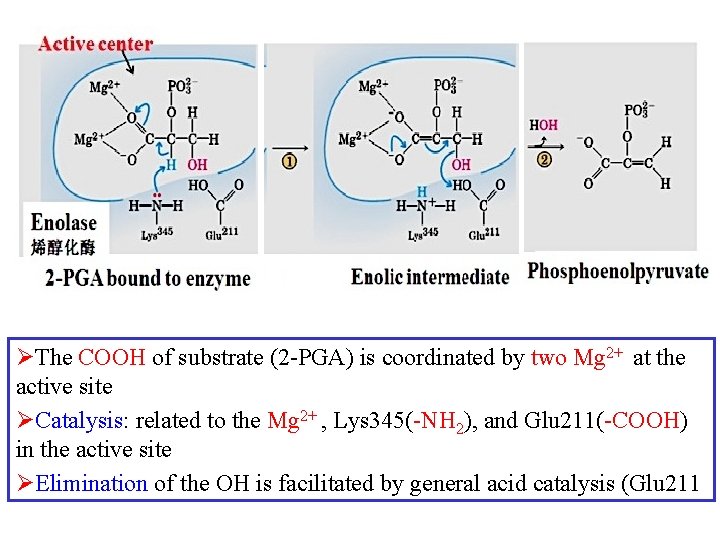
ØThe COOH of substrate (2 -PGA) is coordinated by two Mg 2+ at the active site ØCatalysis: related to the Mg 2+ , Lys 345(-NH 2), and Glu 211(-COOH) in the active site ØElimination of the OH is facilitated by general acid catalysis (Glu 211
Summary Ø E-catalyzed reactions are characterized as high efficiency, high specificity, and regulatory E activity Ø E accelerates the rates of reactions by lowing activation energy Ø Binding of S to the active site of an E which facilitates ES formation requires many reactions including acidbase, metal ion, nucleophilic catalysis…

Section III Enzyme kinetics 酶促反�� 力学 Ø Michaelis-Menten equation(米氏方程)* Ø Km and Vmax*** Ø Effect of p. H, T on velocity of reaction***
![Enzyme kinetics Study of factors which affect the rate of enzymecatalyzed reactions: Ø [S] Enzyme kinetics Study of factors which affect the rate of enzymecatalyzed reactions: Ø [S]](http://slidetodoc.com/presentation_image_h2/75cf664e19fed31f308bfa56af7eb149/image-63.jpg)
Enzyme kinetics Study of factors which affect the rate of enzymecatalyzed reactions: Ø [S] (底物浓度) Ø [E] (酶的浓度) Ø p. H, temperature Ø inhibitor and activator 抑制剂和激活剂 Ø Describing of an enzyme affinity (亲和力) for a substrate
![Michaelis-Menten equation* 米-曼氏方程 ---Study effect of [S] on velocity Hypothesis: !Single substrate, one product Michaelis-Menten equation* 米-曼氏方程 ---Study effect of [S] on velocity Hypothesis: !Single substrate, one product](http://slidetodoc.com/presentation_image_h2/75cf664e19fed31f308bfa56af7eb149/image-65.jpg)
Michaelis-Menten equation* 米-曼氏方程 ---Study effect of [S] on velocity Hypothesis: !Single substrate, one product ! Formation of the ES complex ! [S]>>[E] The M-M equation was proposed in 1913
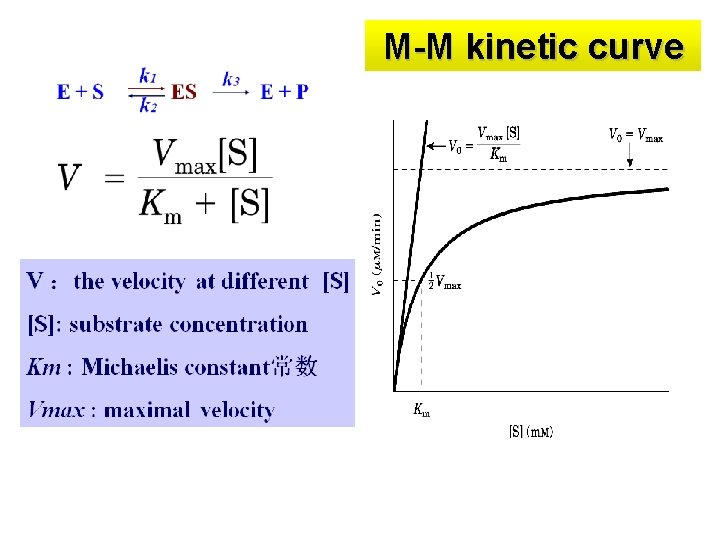
M-M kinetic curve
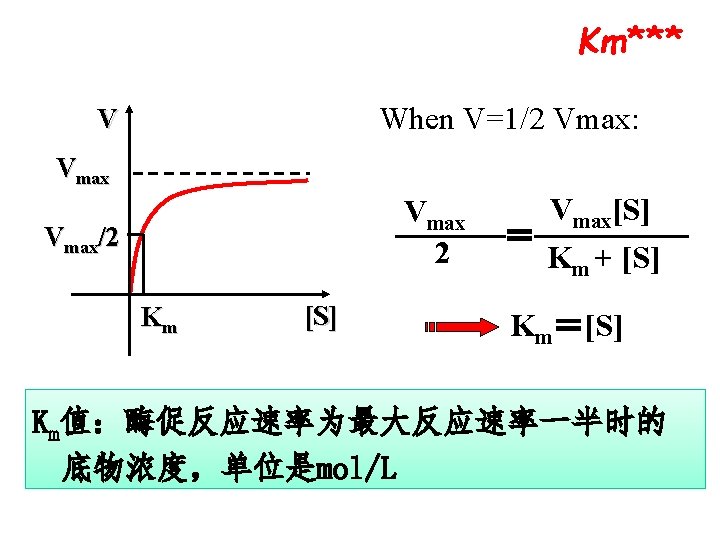
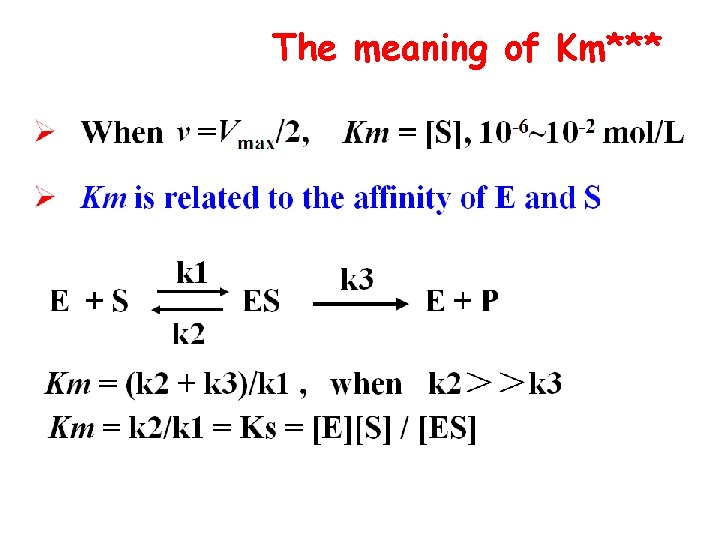
The meaning of Km***
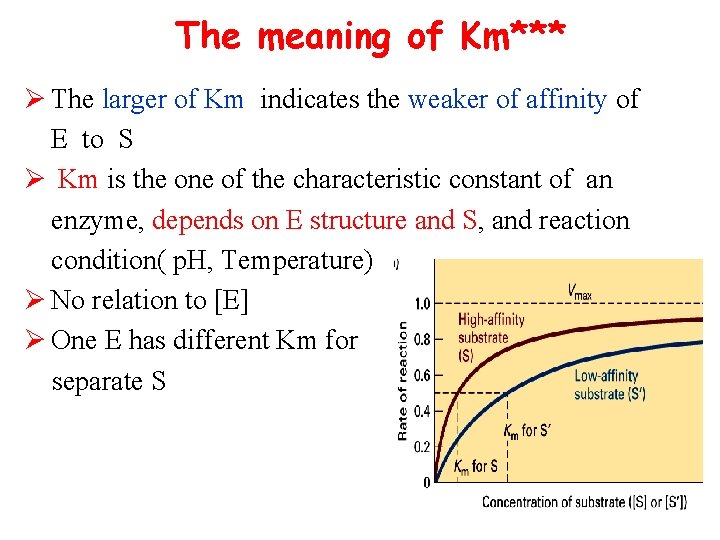
The meaning of Km*** Ø The larger of Km indicates the weaker of affinity of E to S Ø Km is the one of the characteristic constant of an enzyme, depends on E structure and S, and reaction condition( p. H, Temperature) Ø No relation to [E] Ø One E has different Km for separate S
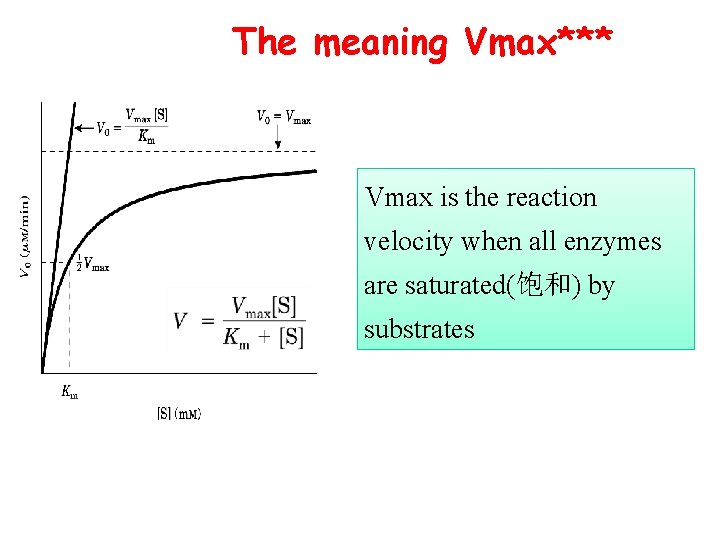
The meaning Vmax*** Vmax is the reaction velocity when all enzymes are saturated(饱和) by substrates
Measurement of Vmax and Km Ø Determination of Km from curve plots is not accurate Ø Double reciprocal plot(Lineweaver--Burk plot, 林—贝氏双倒数法) Ø Hanes-Woolf plot
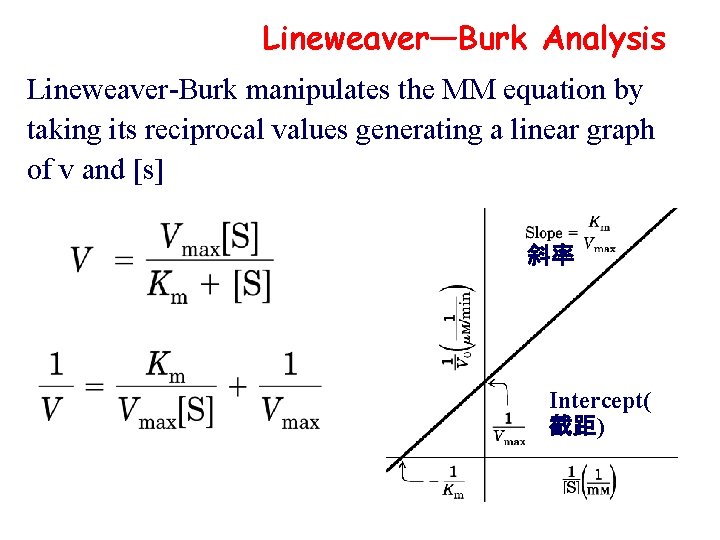
Lineweaver—Burk Analysis Lineweaver-Burk manipulates the MM equation by taking its reciprocal values generating a linear graph of v and [s] 斜率 Intercept( 截距)
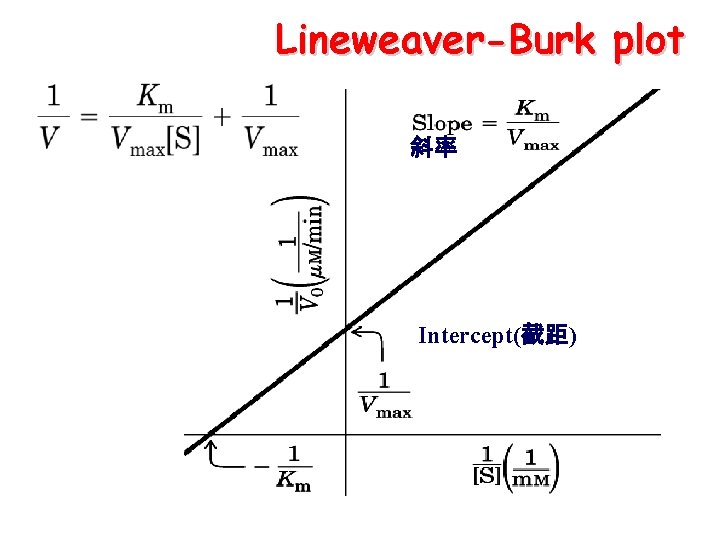
Lineweaver-Burk plot 斜率 Intercept(截距)
![Hanes analysis 在林-� 氏方程基� 上,两� 同乘[S] [S]/V=Km/Vmax + [S]/Vmax Km/Vm -Km [S] Hanes analysis 在林-� 氏方程基� 上,两� 同乘[S] [S]/V=Km/Vmax + [S]/Vmax Km/Vm -Km [S]](http://slidetodoc.com/presentation_image_h2/75cf664e19fed31f308bfa56af7eb149/image-74.jpg)
Hanes analysis 在林-� 氏方程基� 上,两� 同乘[S] [S]/V=Km/Vmax + [S]/Vmax Km/Vm -Km [S]
![Michaelis-Menten equation* 米-曼氏方程 ---Study effect of [S] on velocity Ø Effects of [S] on Michaelis-Menten equation* 米-曼氏方程 ---Study effect of [S] on velocity Ø Effects of [S] on](http://slidetodoc.com/presentation_image_h2/75cf664e19fed31f308bfa56af7eb149/image-75.jpg)
Michaelis-Menten equation* 米-曼氏方程 ---Study effect of [S] on velocity Ø Effects of [S] on velocity Ø Quantitatively(定量) describes the relationship between the V and [S]
Factors Affecting Enzyme Activity Ø Substrate concentration—MM equation Ø Enzyme concentration Ø Temperature, p. H Ø Inhibitors Ø Activators
![Effect of [E] on velocity V When S is enough [S]>>[E], V = K Effect of [E] on velocity V When S is enough [S]>>[E], V = K](http://slidetodoc.com/presentation_image_h2/75cf664e19fed31f308bfa56af7eb149/image-77.jpg)
Effect of [E] on velocity V When S is enough [S]>>[E], V = K 3 [E] 0 [E]
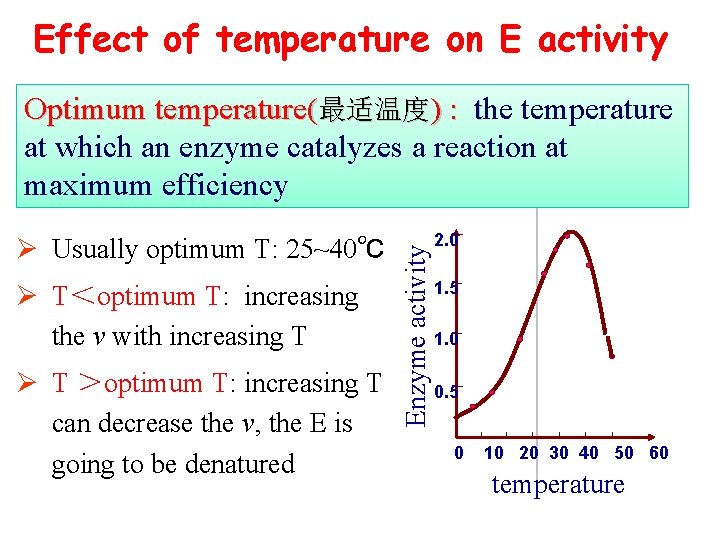
Effect of temperature on E activity Ø Usually optimum T: 25~40℃ Ø T<optimum T: increasing the v with increasing T Ø T >optimum T: increasing T can decrease the v, the E is going to be denatured Enzyme activity Optimum temperature(最适温度) : the temperature at which an enzyme catalyzes a reaction at maximum efficiency 2. 0 1. 5 1. 0 0. 5 0 10 20 30 40 50 60 temperature
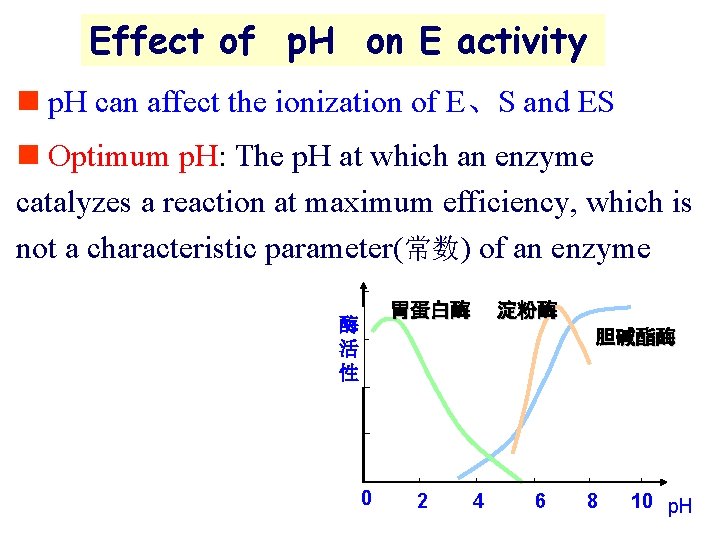
Effect of p. H on E activity n p. H can affect the ionization of E、S and ES n Optimum p. H: The p. H at which an enzyme catalyzes a reaction at maximum efficiency, which is not a characteristic parameter(常数) of an enzyme 胃蛋白酶 酶 活 性 淀粉酶 胆碱酯酶 0 2 4 6 8 10 p. H
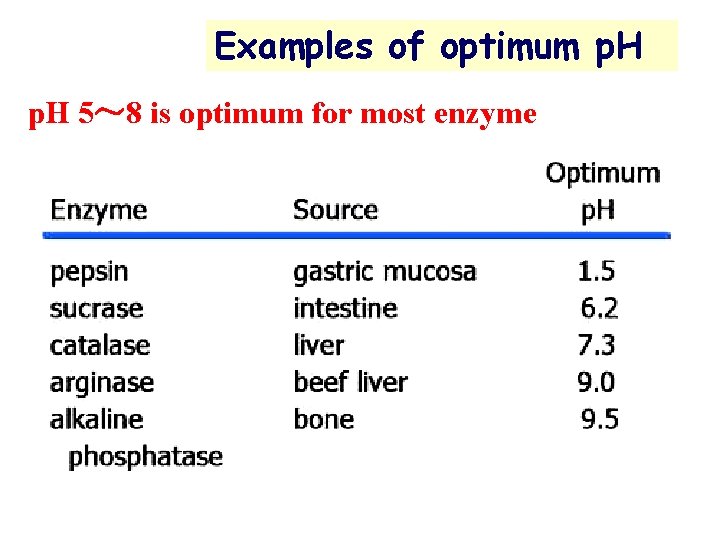
Examples of optimum p. H 5~ 8 is optimum for most enzyme
- Slides: 80