Chapter 3 DNA RNA and Protein Synthesis Processing

- Slides: 17

Chapter 3. DNA, RNA, and Protein Synthesis

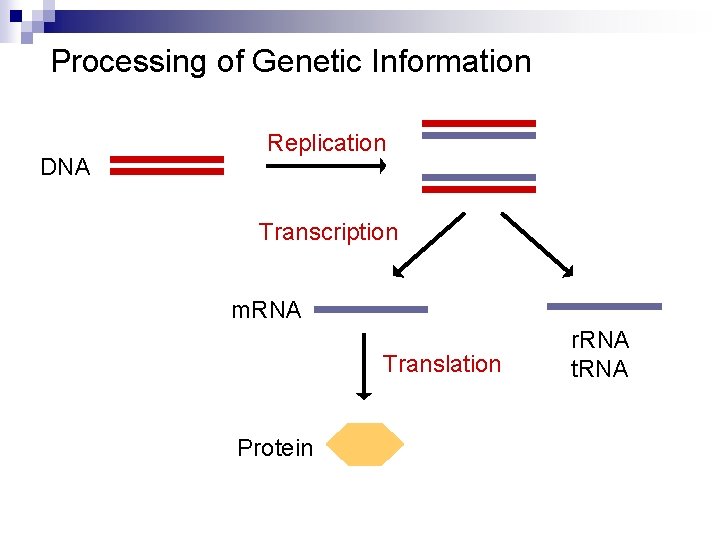
Processing of Genetic Information DNA Replication Transcription m. RNA Translation Protein r. RNA t. RNA

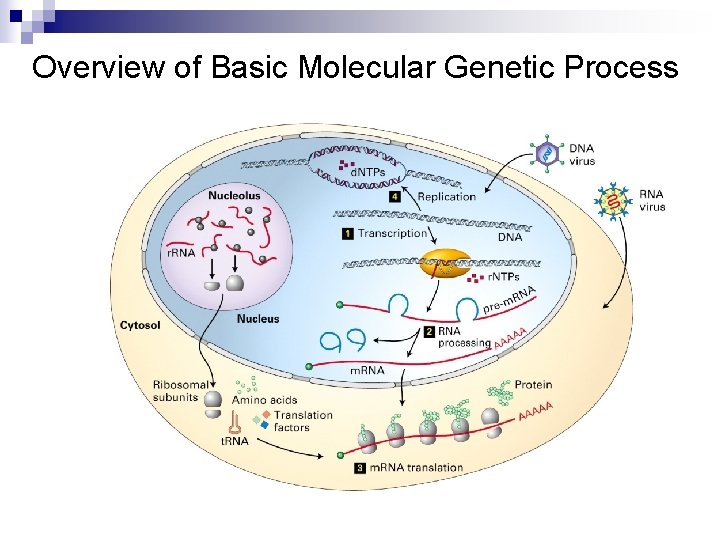
Overview of Basic Molecular Genetic Process

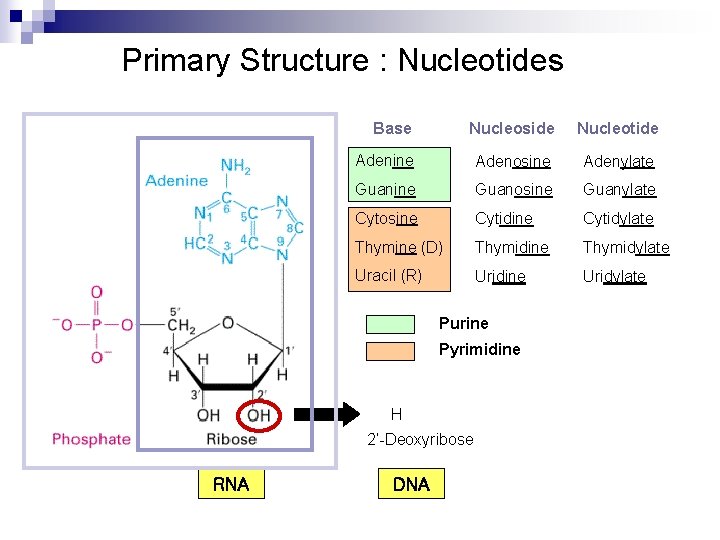
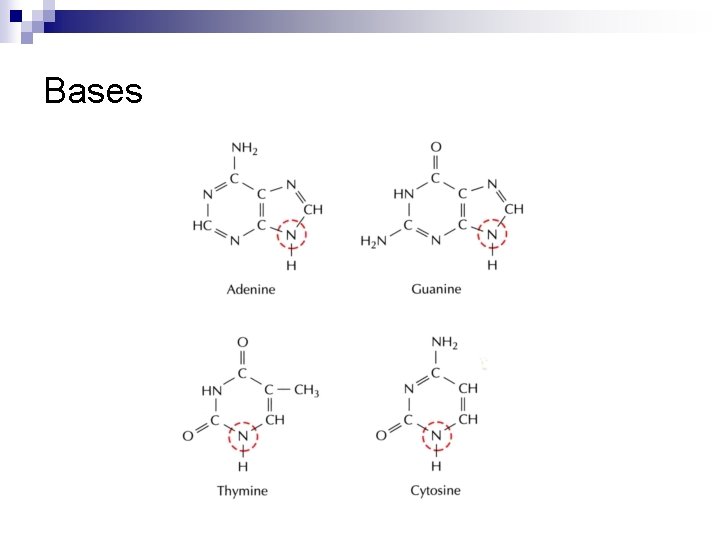
Primary Structure : Nucleotides Base Nucleoside Nucleotide Adenine Adenosine Adenylate Guanine Guanosine Guanylate Cytosine Cytidylate Thymine (D) Thymidine Thymidylate Uracil (R) Uridine Uridylate Purine Pyrimidine H 2’-Deoxyribose RNA DNA

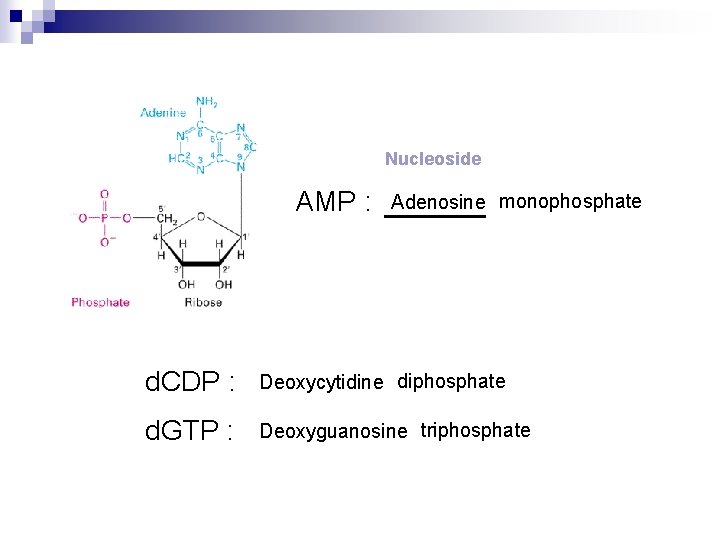
Nucleoside AMP : Adenosine monophosphate d. CDP : Deoxycytidine diphosphate d. GTP : Deoxyguanosine triphosphate

Bases

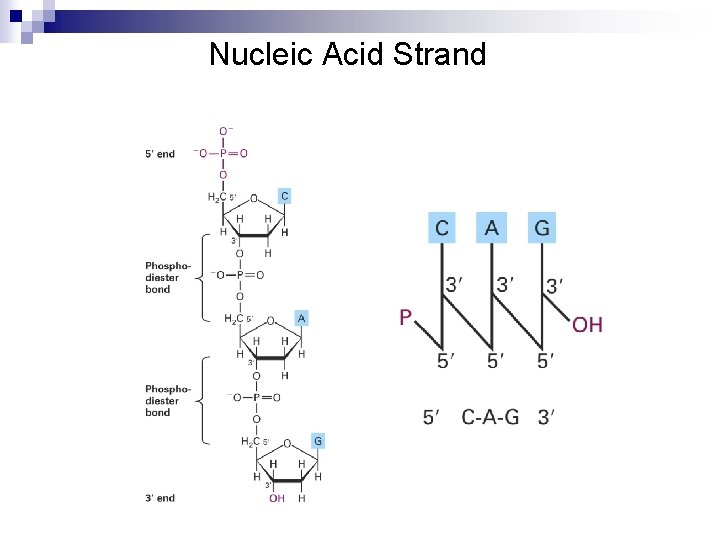
Nucleic Acid Strand

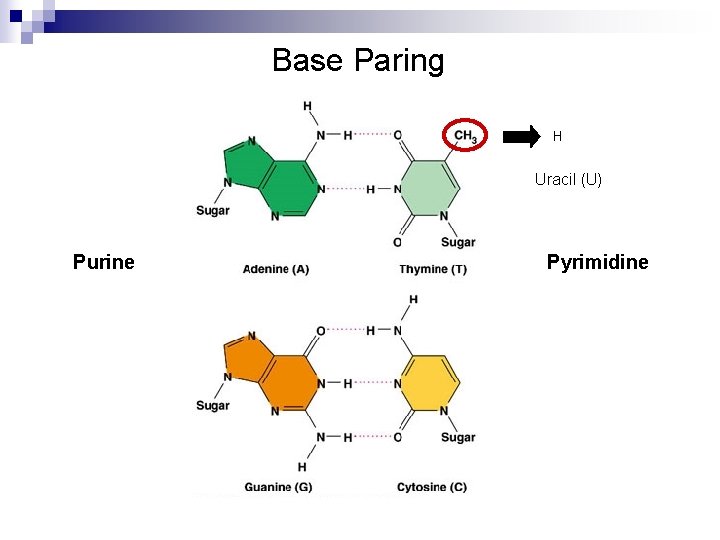
DNA Double Helix n DNA double helix structure ¨ Two antiparallel chains of deoxyribose-phosphate backbone ¨ Two chains are joined by complementary base pairing n n n A: : T, G: : : C Hydrogen bonding DNA size ¨ Indicated by the number of base pairs ¨ Kb: 103 bp, Mb: 106 bp

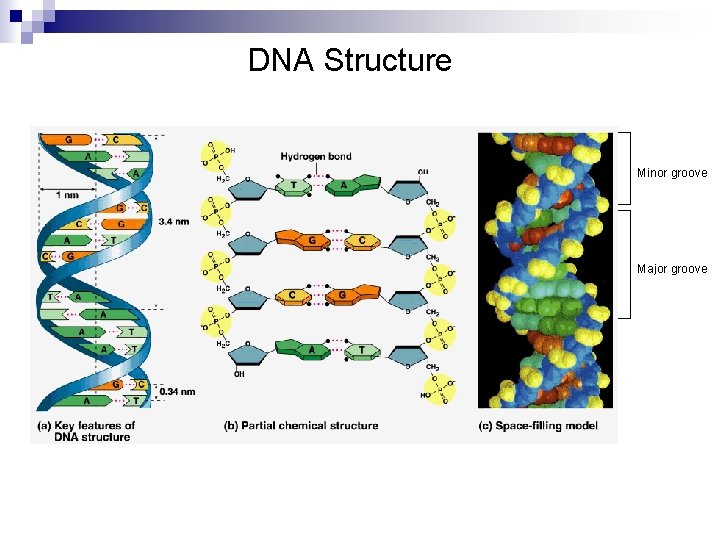
DNA Structure Minor groove Major groove

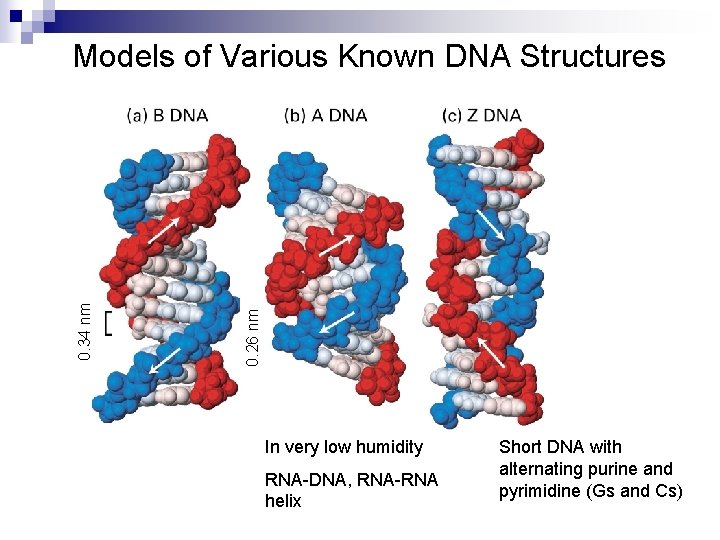
0. 26 nm 0. 34 nm Models of Various Known DNA Structures In very low humidity RNA-DNA, RNA-RNA helix Short DNA with alternating purine and pyrimidine (Gs and Cs)

Base Paring H Uracil (U) Purine Pyrimidine

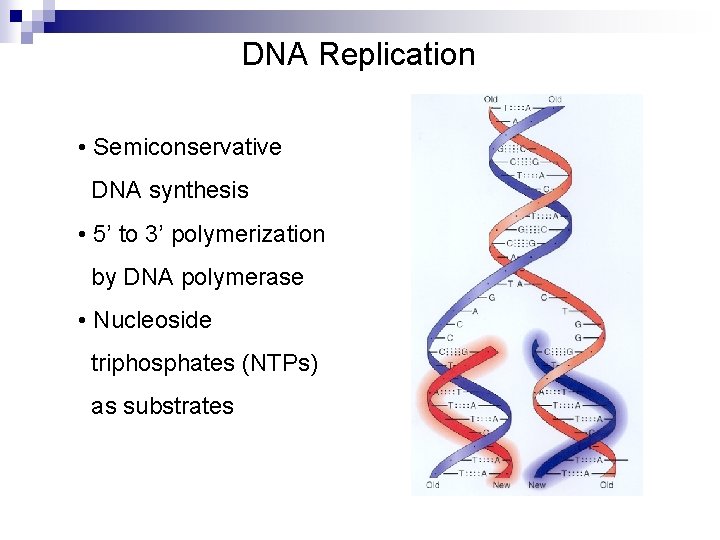
DNA Replication • Semiconservative DNA synthesis • 5’ to 3’ polymerization by DNA polymerase • Nucleoside triphosphates (NTPs) as substrates

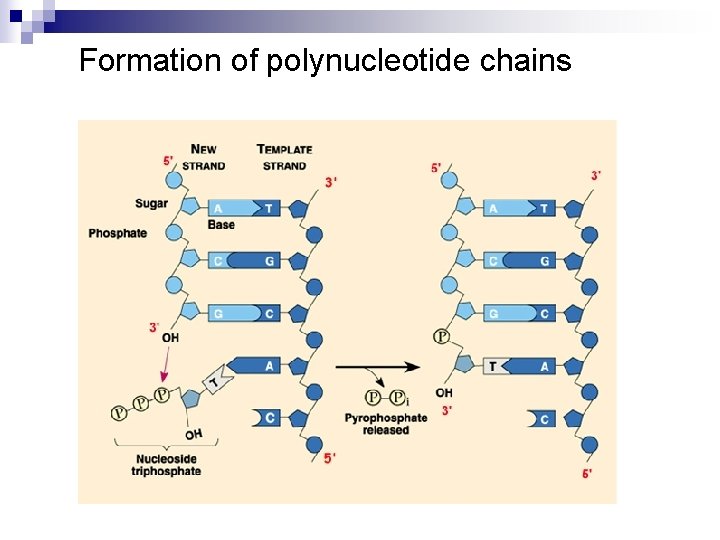
Formation of polynucleotide chains

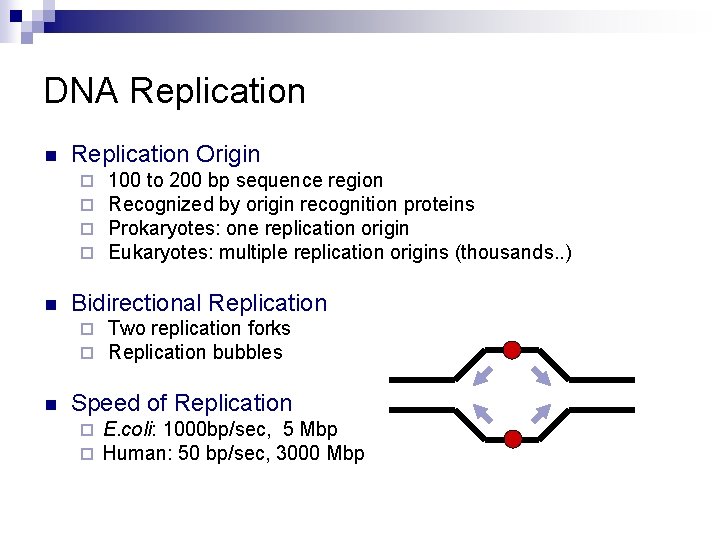
DNA Replication n Replication Origin ¨ ¨ n Bidirectional Replication ¨ ¨ n 100 to 200 bp sequence region Recognized by origin recognition proteins Prokaryotes: one replication origin Eukaryotes: multiple replication origins (thousands. . ) Two replication forks Replication bubbles Speed of Replication ¨ ¨ E. coli: 1000 bp/sec, 5 Mbp Human: 50 bp/sec, 3000 Mbp

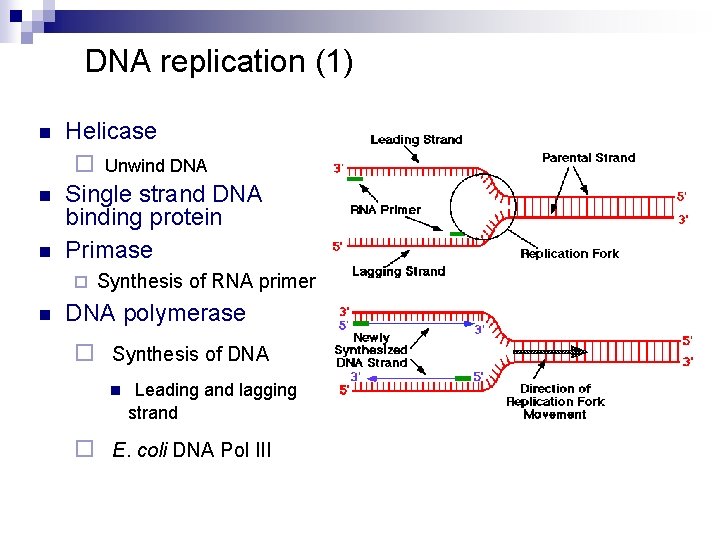
DNA replication (1) n n n Helicase ¨ Unwind DNA Single strand DNA binding protein Primase ¨ n Synthesis of RNA primer DNA polymerase ¨ Synthesis of DNA n Leading and lagging strand ¨ E. coli DNA Pol III

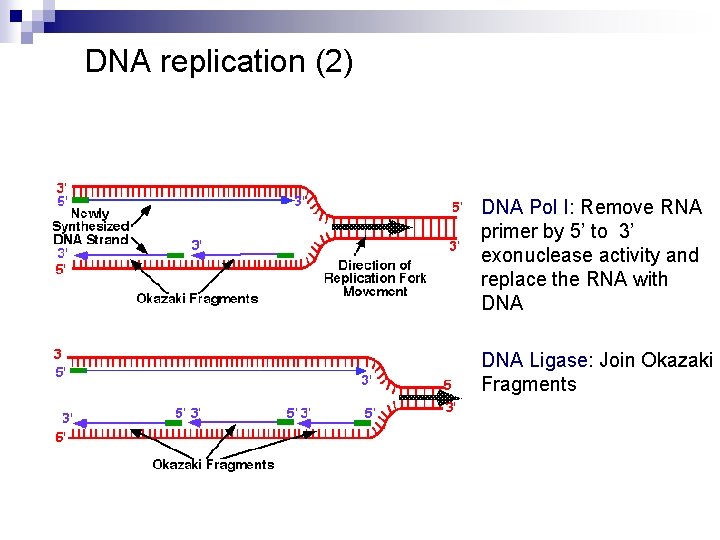
DNA replication (2) DNA Pol I: Remove RNA primer by 5’ to 3’ exonuclease activity and replace the RNA with DNA Ligase: Join Okazaki Fragments

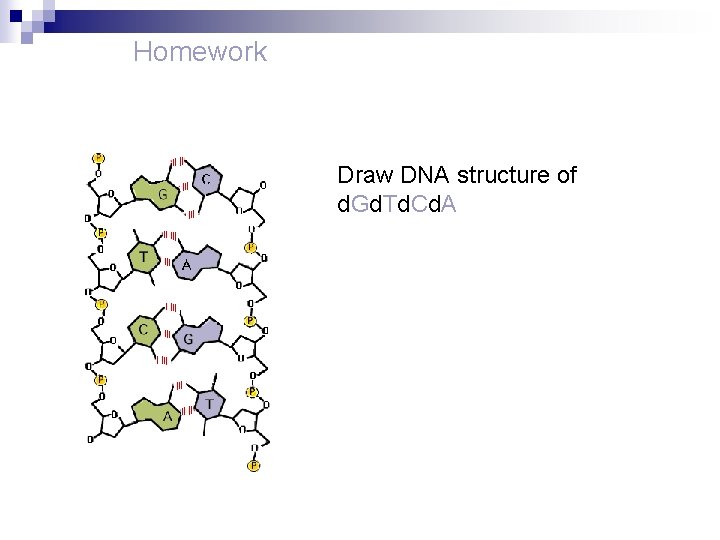
Homework Draw DNA structure of d. Gd. Td. Cd. A