Chapter 2 Bayesian hierarchical models in geographical genetics

Chapter 2: Bayesian hierarchical models in geographical genetics Manda Sayler
• Geographical genetics is the field of population genetics that focuses on describing the distribution of genetic variation within and among populations and understanding the processes that produce those patterns. • Statistical sampling uncertainty arises from the process of constructing allele frequency estimates from population samples. • Genetic sampling uncertainty arises from the underlying stochastic evolutionary process that gave rise to the population we sampled. – Note: increasing the sample size of alleles with each population reduces statistical uncertainty, but it cannot reduce the magnitude of genetic uncertainty. • Weir and Cockerham approach is the most widely used approach for analysis of genetic diversity in hierarchically structured populations. • Bayesian approach provides a model-based approach to inference that is enormously powerful and flexible. • Hierarchical Bayesian models provide a natural approach to inference in geographical genetics.
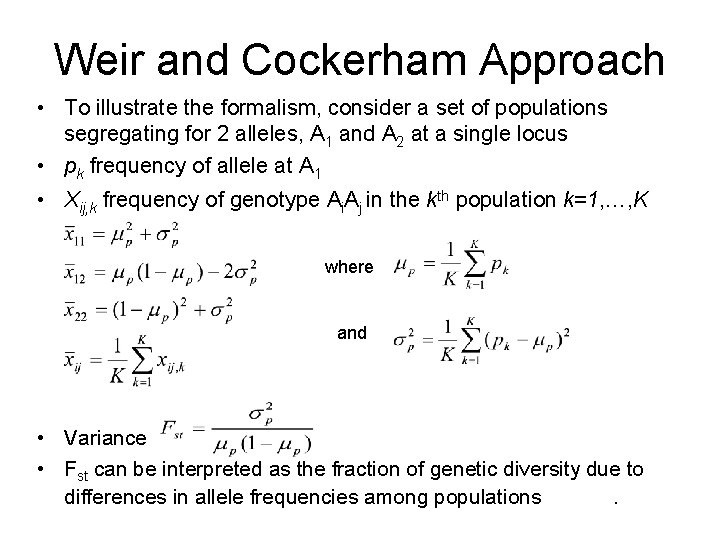
Weir and Cockerham Approach • To illustrate the formalism, consider a set of populations segregating for 2 alleles, A 1 and A 2 at a single locus • pk frequency of allele at A 1 • Xij, k frequency of genotype Ai. Aj in the kth population k=1, …, K where and • Variance • Fst can be interpreted as the fraction of genetic diversity due to differences in allele frequencies among populations.
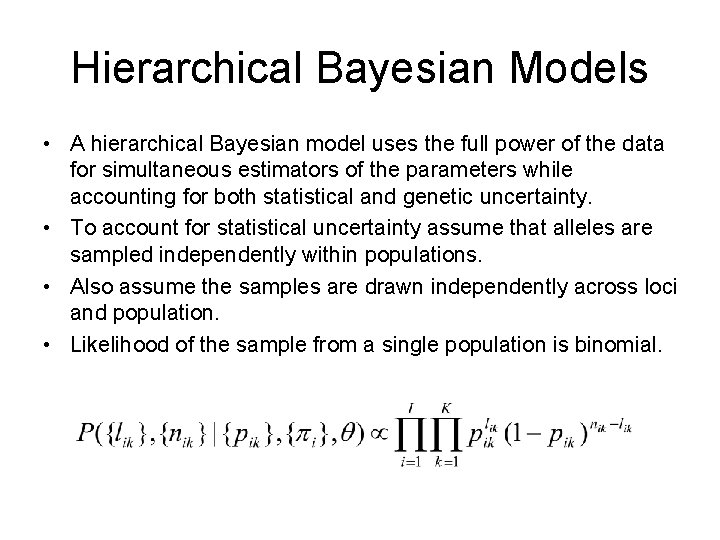
Hierarchical Bayesian Models • A hierarchical Bayesian model uses the full power of the data for simultaneous estimators of the parameters while accounting for both statistical and genetic uncertainty. • To account for statistical uncertainty assume that alleles are sampled independently within populations. • Also assume the samples are drawn independently across loci and population. • Likelihood of the sample from a single population is binomial.
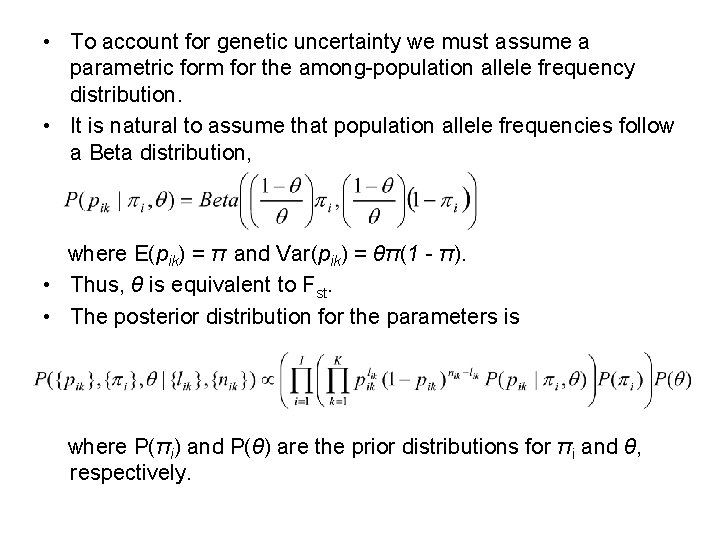
• To account for genetic uncertainty we must assume a parametric form for the among-population allele frequency distribution. • It is natural to assume that population allele frequencies follow a Beta distribution, where E(pik) = π and Var(pik) = θπ(1 - π). • Thus, θ is equivalent to Fst. • The posterior distribution for the parameters is where P(πi) and P(θ) are the prior distributions for πi and θ, respectively.
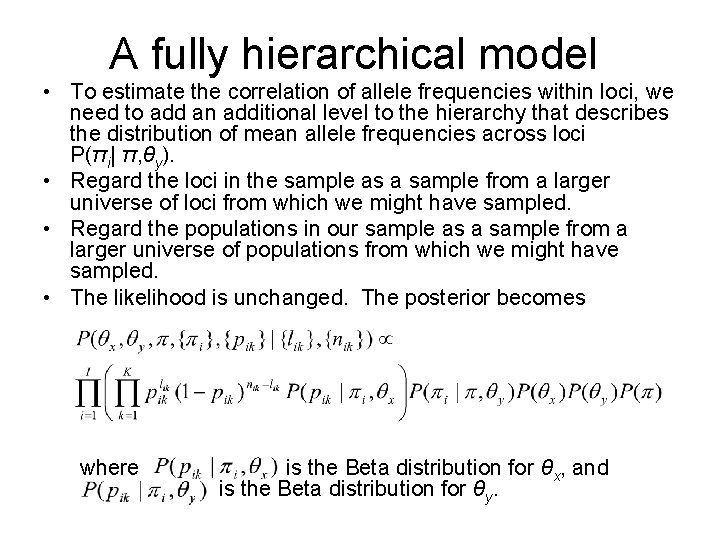
A fully hierarchical model • To estimate the correlation of allele frequencies within loci, we need to add an additional level to the hierarchy that describes the distribution of mean allele frequencies across loci P(πi| π, θy). • Regard the loci in the sample as a sample from a larger universe of loci from which we might have sampled. • Regard the populations in our sample as a sample from a larger universe of populations from which we might have sampled. • The likelihood is unchanged. The posterior becomes where is the Beta distribution for θx, and is the Beta distribution for θy.
Developing an MCMC sampler • The process begins by picking an initial value for p, called p 0, then p 0 is updated until we have a large sample of values pt using either – Metropolis-Hastings algorithm (Figure 2. 2) – Slice algorithm (Figure 2. 3) • Estimate any property of the posterior to an arbitrary degree of accuracy. • Ensure that the MC has converged the values from an initial burn-in period are discarded. • Values retained from the following sample period represent the full posterior distribution and summary statistics are calculated directly from this sample. • Reduce the autocorrelation of values in the sample, it is sometimes useful to thin the sample.
- Slides: 7