Chapter 16 Variations in Chromosome Structure and Function







- Slides: 40

Chapter 16 - Variations in Chromosome Structure and Function: • • Chromosome structure • Deletion, duplication, inversion, translocation • Focus of Cytogenetics • Source of new genes with new functions • Reproductive isolation mechanism Chromosome number • Aneuploidy, monoploidy, and polyploidy • Can lead to speciation

Chromosomal mutations: • Arise spontaneously or can be induced by chemicals or radiation. • Major contributors to human miscarriage, stillbirths, and genetic disorders. • ~1/2 of spontaneous abortions result from chromosomal mutations. • Visible (microscope) mutations occur in 6/1, 000 live births. • ~11% of men with fertility problems and 6% of men with mental deficiencies possess chromosomal mutations.

Chromosomal structure mutations: 1. Deletion 2. Duplication 3. Inversion - changing orientation of a DNA segment 4. Translocation - moving a DNA segment

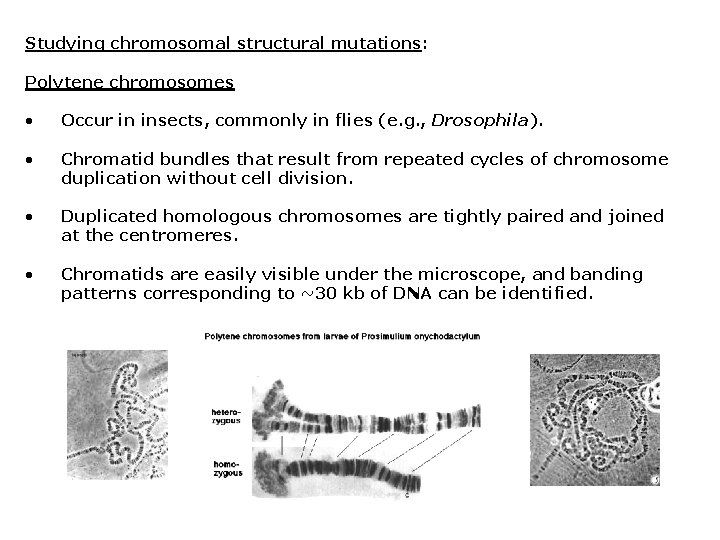
Studying chromosomal structural mutations: Polytene chromosomes • Occur in insects, commonly in flies (e. g. , Drosophila). • Chromatid bundles that result from repeated cycles of chromosome duplication without cell division. • Duplicated homologous chromosomes are tightly paired and joined at the centromeres. • Chromatids are easily visible under the microscope, and banding patterns corresponding to ~30 kb of DNA can be identified.

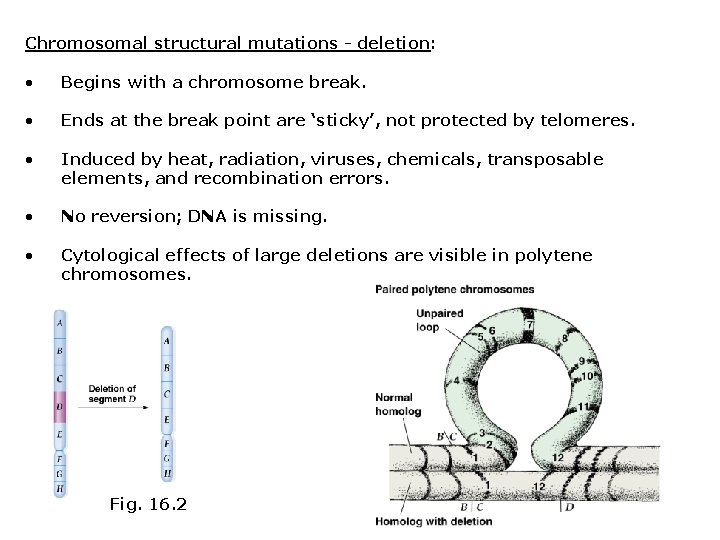
Chromosomal structural mutations - deletion: • Begins with a chromosome break. • Ends at the break point are ‘sticky’, not protected by telomeres. • Induced by heat, radiation, viruses, chemicals, transposable elements, and recombination errors. • No reversion; DNA is missing. • Cytological effects of large deletions are visible in polytene chromosomes. Fig. 16. 2

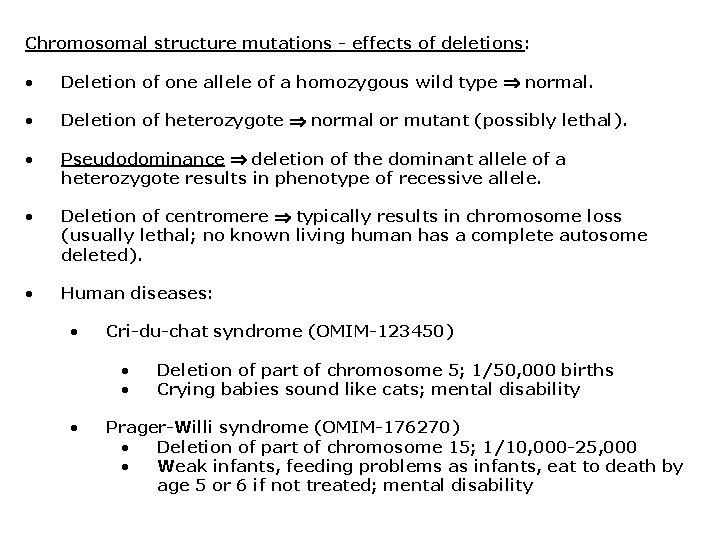
Chromosomal structure mutations - effects of deletions: • Deletion of one allele of a homozygous wild type normal. • Deletion of heterozygote normal or mutant (possibly lethal). • Pseudodominance deletion of the dominant allele of a heterozygote results in phenotype of recessive allele. • Deletion of centromere typically results in chromosome loss (usually lethal; no known living human has a complete autosome deleted). • Human diseases: • Cri-du-chat syndrome (OMIM-123450) • • • Deletion of part of chromosome 5; 1/50, 000 births Crying babies sound like cats; mental disability Prager-Willi syndrome (OMIM-176270) • Deletion of part of chromosome 15; 1/10, 000 -25, 000 • Weak infants, feeding problems as infants, eat to death by age 5 or 6 if not treated; mental disability

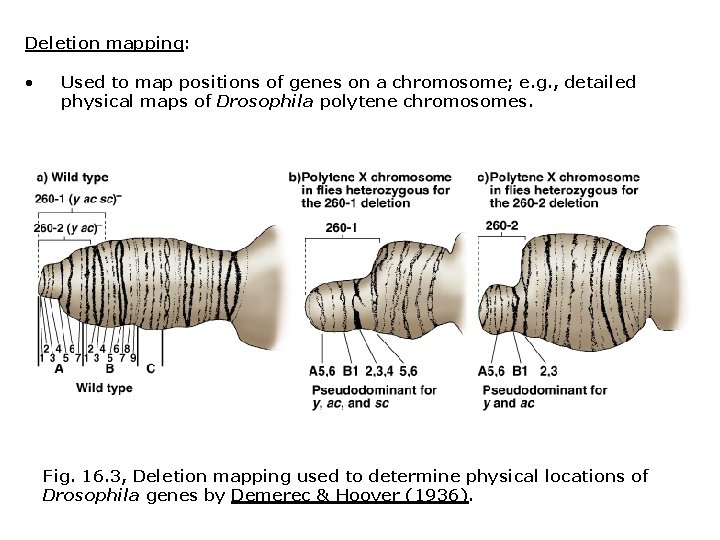
Deletion mapping: • Used to map positions of genes on a chromosome; e. g. , detailed physical maps of Drosophila polytene chromosomes. Fig. 16. 3, Deletion mapping used to determine physical locations of Drosophila genes by Demerec & Hoover (1936).

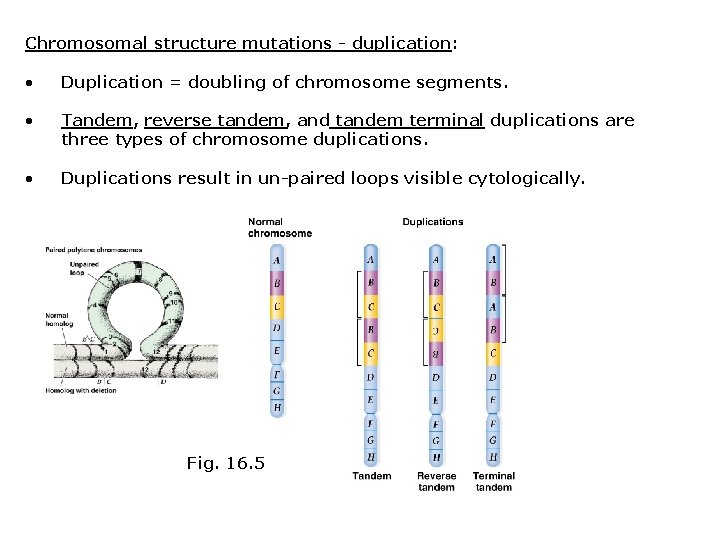
Chromosomal structure mutations - duplication: • Duplication = doubling of chromosome segments. • Tandem, reverse tandem, and tandem terminal duplications are three types of chromosome duplications. • Duplications result in un-paired loops visible cytologically. Fig. 16. 5

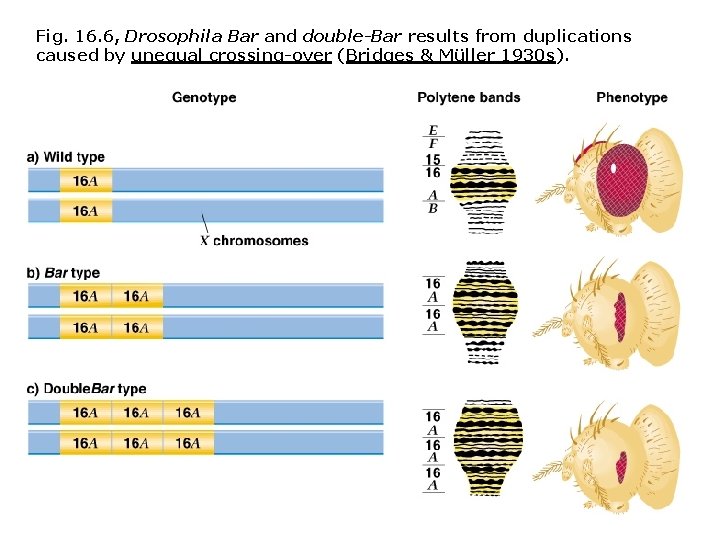
Fig. 16. 6, Drosophila Bar and double-Bar results from duplications caused by unequal crossing-over (Bridges & Müller 1930 s).

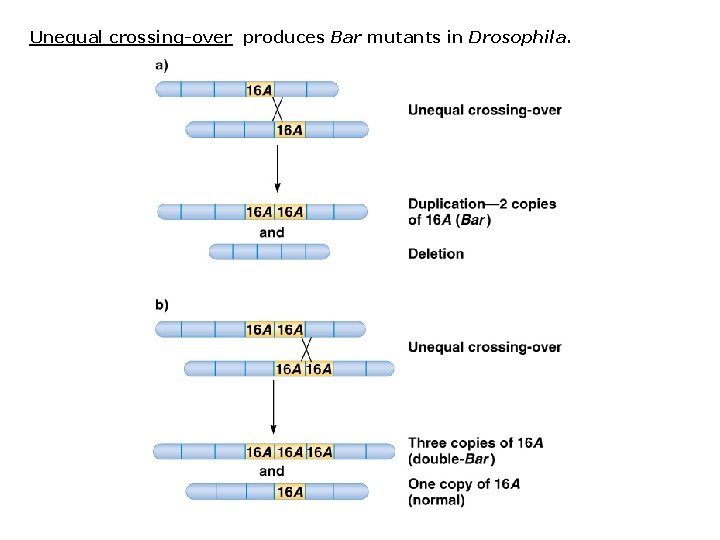
Unequal crossing-over produces Bar mutants in Drosophila.

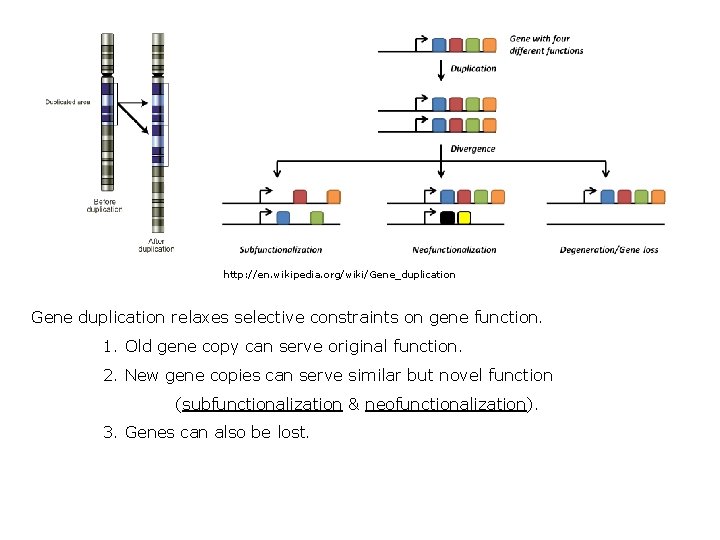
http: //en. wikipedia. org/wiki/Gene_duplication Gene duplication relaxes selective constraints on gene function. 1. Old gene copy can serve original function. 2. New gene copies can serve similar but novel function (subfunctionalization & neofunctionalization). 3. Genes can also be lost.

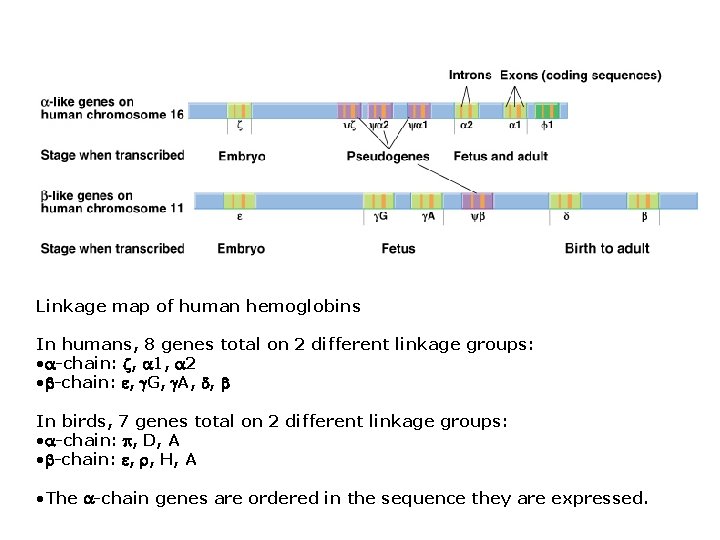
Multi-gene families - result from duplications: Hemoglobins (Hb) • Genes for the -chain are clustered on one chromosome, and genes for the -chain occur on another chromosome. • Each Hb gene contains multiple ORFs; adults and embyros also use different hemoglobins genes. • Adult and embryonic hemoglobins on same chromosomes share similar sequences that arose by duplication. • and hemoglobins also are similar; gene duplication followed by sequence divergence and periodic gene conversion. • Different Hb genes contribute to different isoforms with different biochemical properties (e. g. , fetal vs. adult hemoglobin differ in their affinity for oxygen).

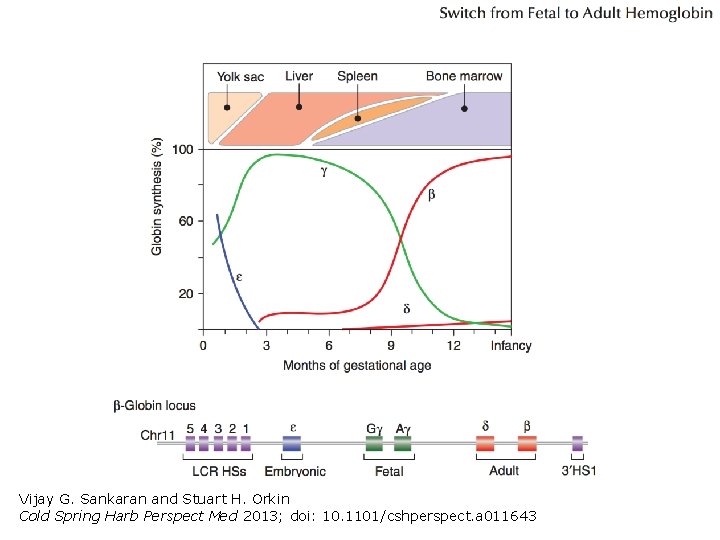
Linkage map of human hemoglobins In humans, 8 genes total on 2 different linkage groups: • -chain: , 1, 2 • -chain: , G, A, , In birds, 7 genes total on 2 different linkage groups: • -chain: , D, A • -chain: , , H, A • The -chain genes are ordered in the sequence they are expressed.

Vijay G. Sankaran and Stuart H. Orkin Cold Spring Harb Perspect Med 2013; doi: 10. 1101/cshperspect. a 011643

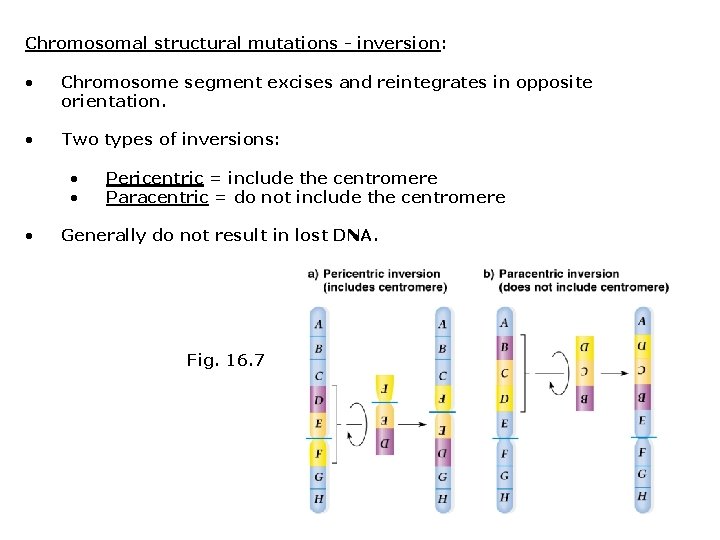
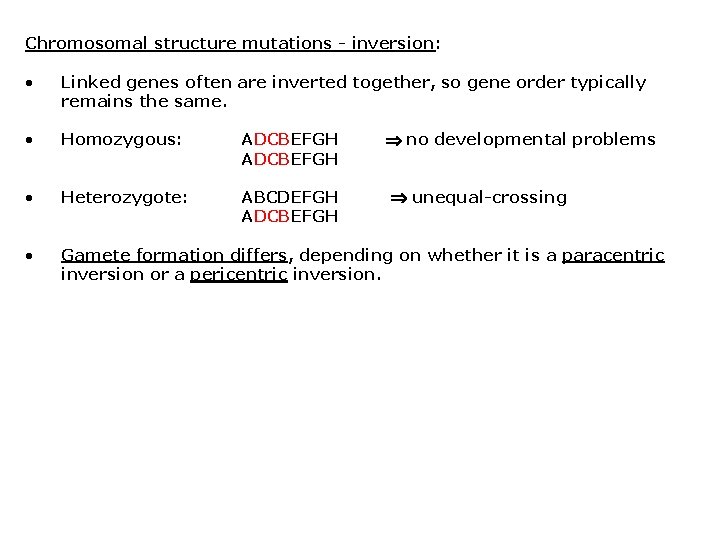
Chromosomal structural mutations - inversion: • Chromosome segment excises and reintegrates in opposite orientation. • Two types of inversions: • • • Pericentric = include the centromere Paracentric = do not include the centromere Generally do not result in lost DNA. Fig. 16. 7

Chromosomal structure mutations - inversion: • Linked genes often are inverted together, so gene order typically remains the same. • Homozygous: ADCBEFGH • Heterozygote: ABCDEFGH ADCBEFGH • Gamete formation differs, depending on whether it is a paracentric inversion or a pericentric inversion. no developmental problems unequal-crossing

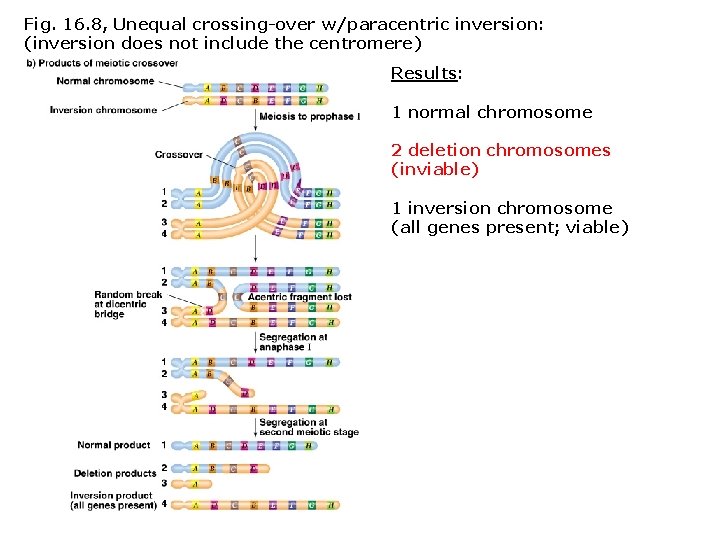
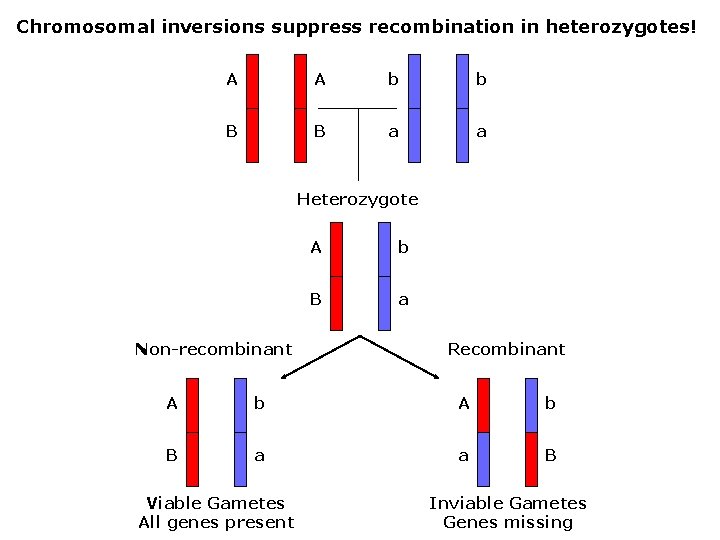
Fig. 16. 8, Unequal crossing-over w/paracentric inversion: (inversion does not include the centromere) Results: 1 normal chromosome 2 deletion chromosomes (inviable) 1 inversion chromosome (all genes present; viable)

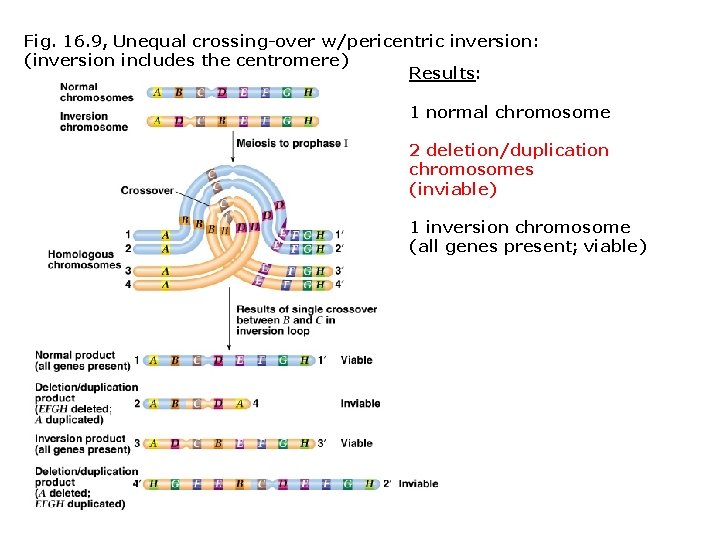
Fig. 16. 9, Unequal crossing-over w/pericentric inversion: (inversion includes the centromere) Results: 1 normal chromosome 2 deletion/duplication chromosomes (inviable) 1 inversion chromosome (all genes present; viable)

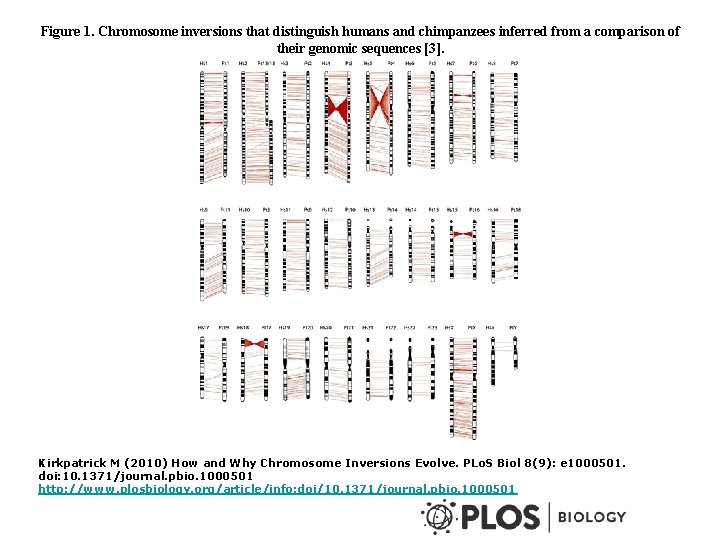
Figure 1. Chromosome inversions that distinguish humans and chimpanzees inferred from a comparison of their genomic sequences [3]. Kirkpatrick M (2010) How and Why Chromosome Inversions Evolve. PLo. S Biol 8(9): e 1000501. doi: 10. 1371/journal. pbio. 1000501 http: //www. plosbiology. org/article/info: doi/10. 1371/journal. pbio. 1000501

Chromosomal inversions suppress recombination in heterozygotes! A A b b B B a a Heterozygote Non-recombinant A b B a Recombinant A b B a a B Viable Gametes All genes present Inviable Gametes Genes missing

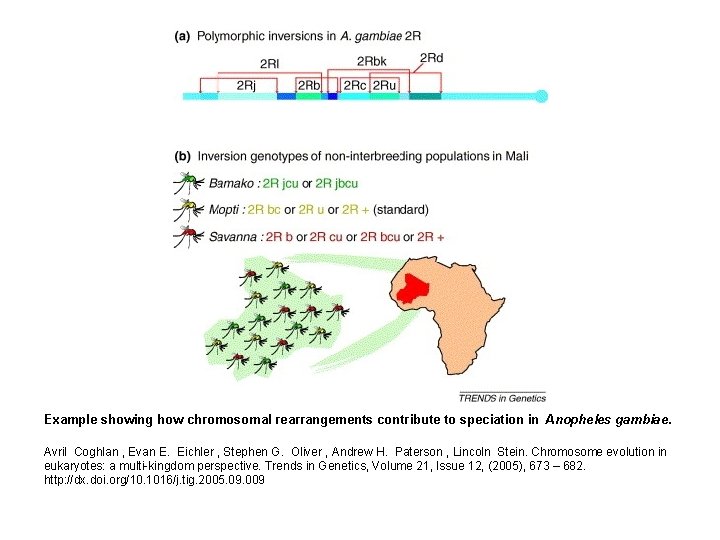
Example showing how chromosomal rearrangements contribute to speciation in Anopheles gambiae. Avril Coghlan , Evan E. Eichler , Stephen G. Oliver , Andrew H. Paterson , Lincoln Stein. Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends in Genetics, Volume 21, Issue 12, (2005), 673 – 682. http: //dx. doi. org/10. 1016/j. tig. 2005. 09. 009

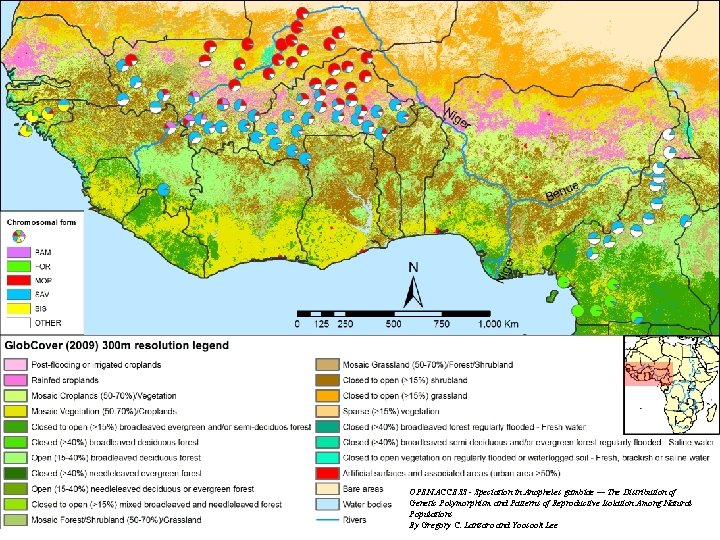
OPEN ACCESS - Speciation in Anopheles gambiae — The Distribution of Genetic Polymorphism and Patterns of Reproductive Isolation Among Natural Populations By Gregory C. Lanzaro and Yoosook Lee

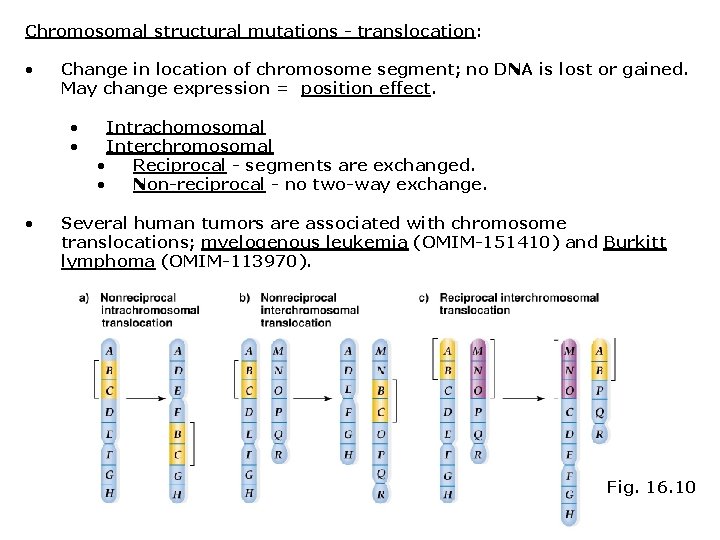
Chromosomal structural mutations - translocation: • Change in location of chromosome segment; no DNA is lost or gained. May change expression = position effect. • • • Intrachomosomal Interchromosomal • Reciprocal - segments are exchanged. • Non-reciprocal - no two-way exchange. Several human tumors are associated with chromosome translocations; myelogenous leukemia (OMIM-151410) and Burkitt lymphoma (OMIM-113970). Fig. 16. 10

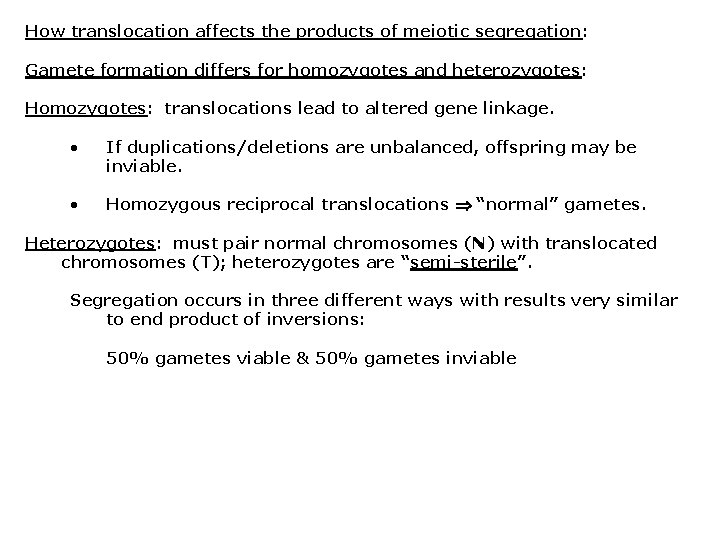
How translocation affects the products of meiotic segregation: Gamete formation differs for homozygotes and heterozygotes: Homozygotes: translocations lead to altered gene linkage. • If duplications/deletions are unbalanced, offspring may be inviable. • Homozygous reciprocal translocations “normal” gametes. Heterozygotes: must pair normal chromosomes (N) with translocated chromosomes (T); heterozygotes are “semi-sterile”. Segregation occurs in three different ways with results very similar to end product of inversions: 50% gametes viable & 50% gametes inviable

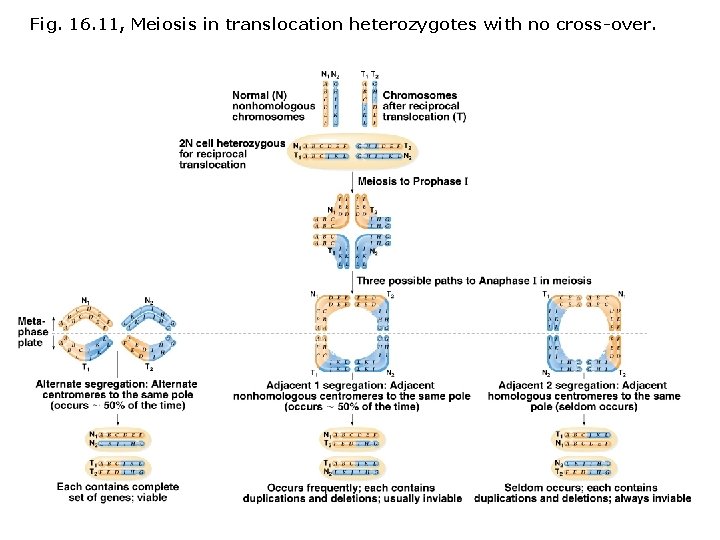
Fig. 16. 11, Meiosis in translocation heterozygotes with no cross-over.

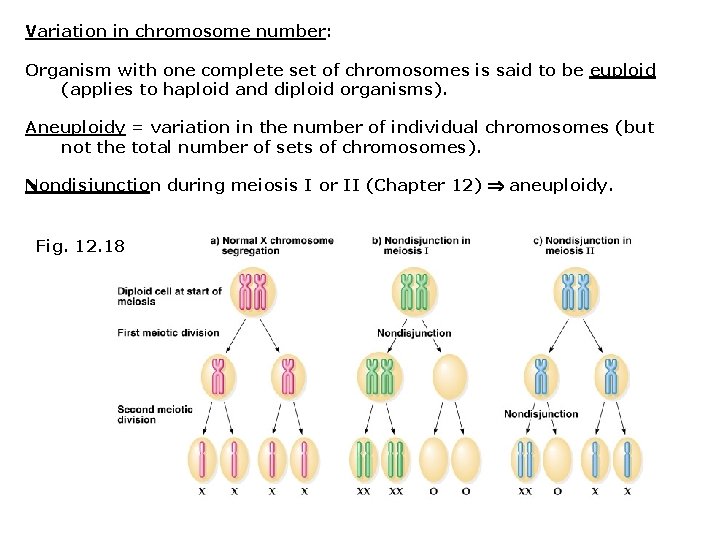
Variation in chromosome number: Organism with one complete set of chromosomes is said to be euploid (applies to haploid and diploid organisms). Aneuploidy = variation in the number of individual chromosomes (but not the total number of sets of chromosomes). Nondisjunction during meiosis I or II (Chapter 12) aneuploidy. Fig. 12. 18

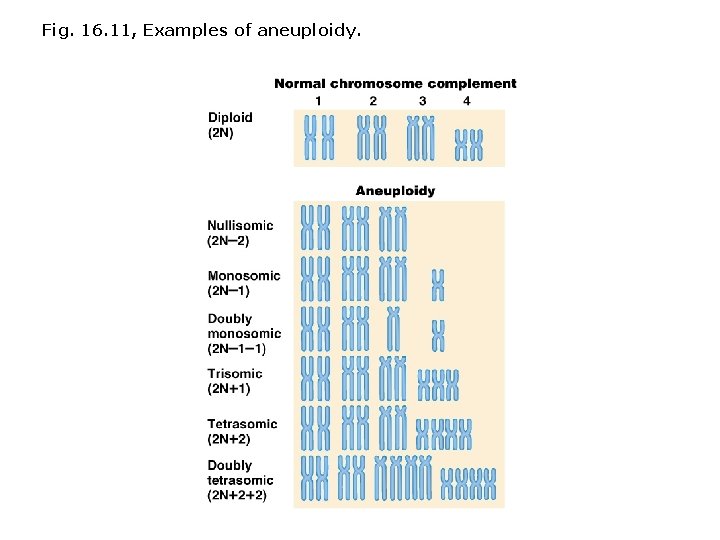
Variation in chromosome number: • Aneuploidy not generally well-tolerated in animals; primarily detected after spontaneous abortion. • Four main types of aneuploidy: Nullisomy = loss of one homologous chromosome pair. Monosomy = loss of a single chromosome. Trisomy = one extra chromosome. Tetrasomy = one extra chromosome pair. • Sex chromosome aneuploidy occurs more often than autosome aneuploidy (inactivation of X compensates). • e. g. , autosomal trisomy accounts for ~1/2 of fetal deaths.

Fig. 16. 11, Examples of aneuploidy.

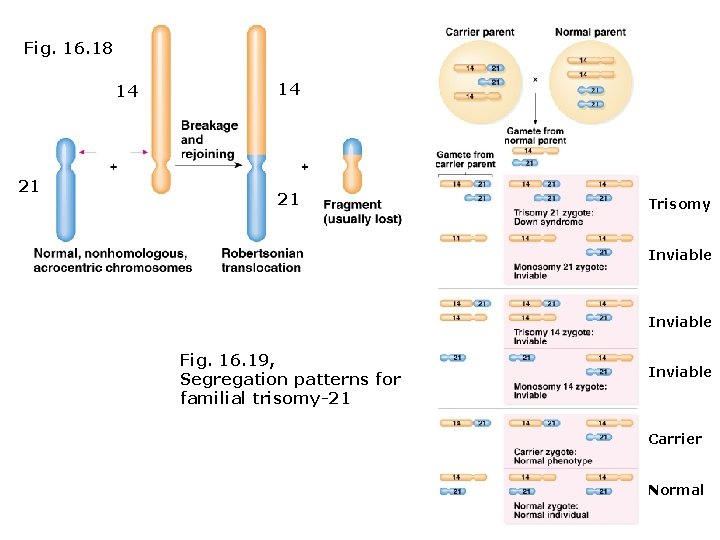
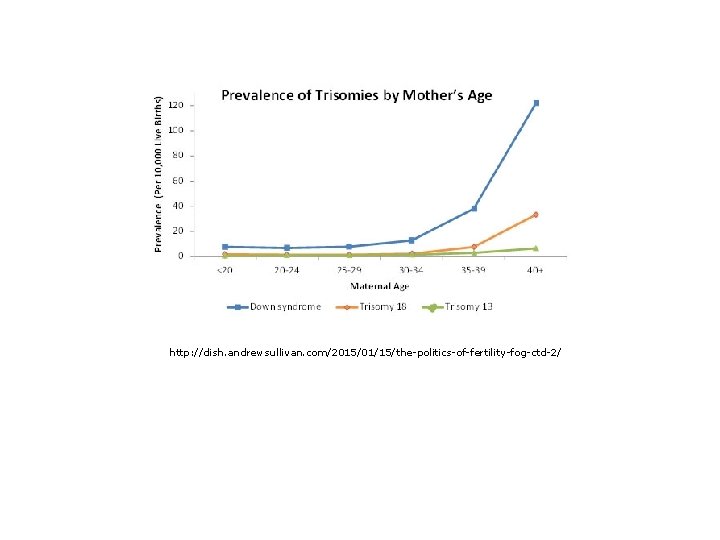
Variation in chromosome number: Down Syndrome (trisomy-21, OMIM-190685): • Occurs in 1/286 conceptions and 1/699 live births. • Probability of non-disjunction trisomy-21 occurring varies with age of ovaries and testes. • Trisomy-21 also occurs by Robertsonian translocation joins long arm of chromosome 21 with long arm of chromosome 14 or 15. • Familial down syndrome arises when carrier parents (heterozygotes) mate with normal parents. • 1/2 gametes are inviable. • 1/3 of live offspring are trisomy-21; 1/3 are carrier heterozygotes, and 1/3 are normal.

Fig. 16. 18 14 21 Trisomy Inviable Fig. 16. 19, Segregation patterns for familial trisomy-21 Inviable Carrier Normal

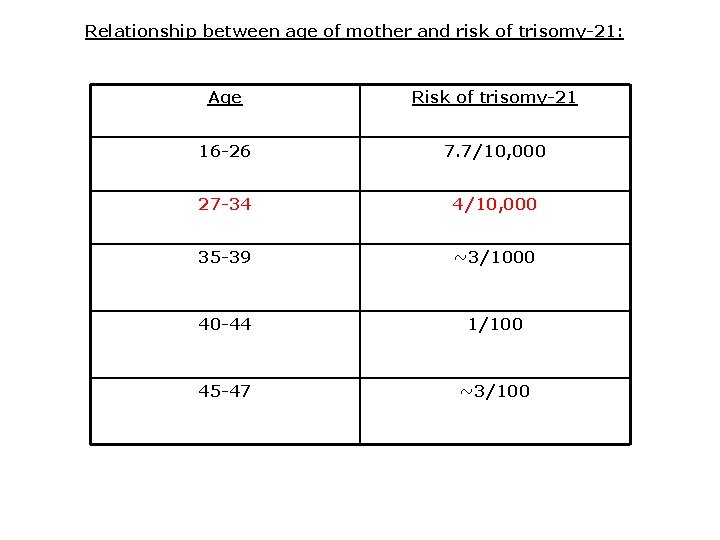
Relationship between age of mother and risk of trisomy-21: Age Risk of trisomy-21 16 -26 7. 7/10, 000 27 -34 4/10, 000 35 -39 ~3/1000 40 -44 1/100 45 -47 ~3/100

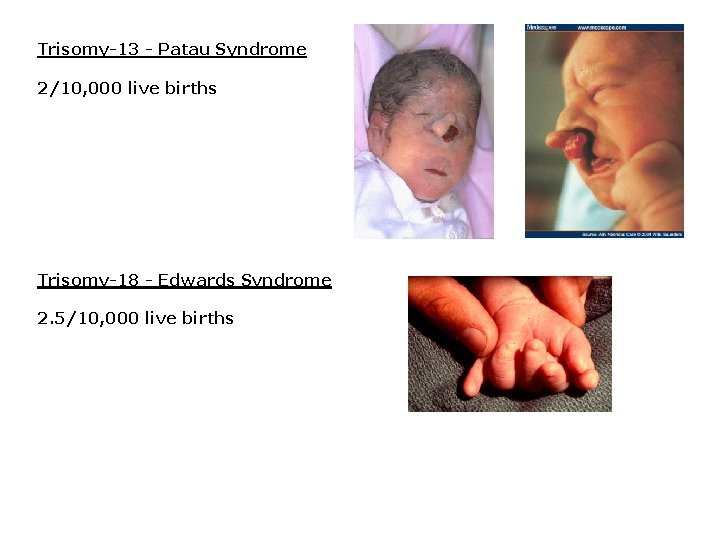
Trisomy-13 - Patau Syndrome 2/10, 000 live births Trisomy-18 - Edwards Syndrome 2. 5/10, 000 live births

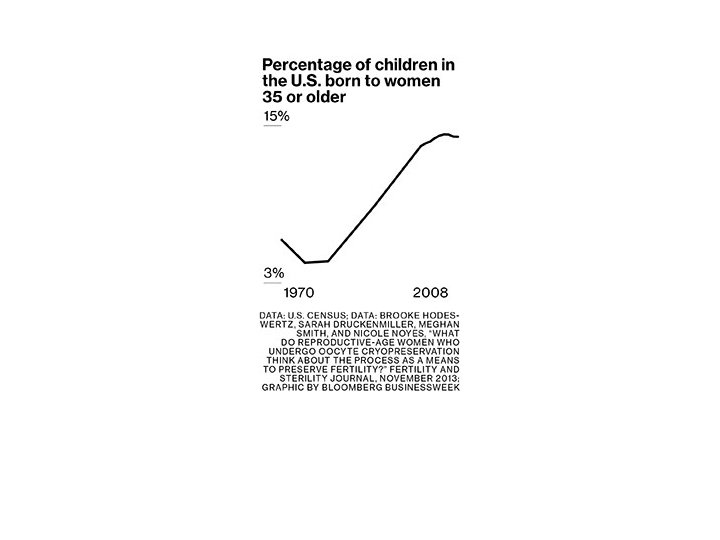
http: //dish. andrewsullivan. com/2015/01/15/the-politics-of-fertility-fog-ctd-2/


http: //food-hacks. wonderhowto. com/how-to/tell-if-your-expired-eggs-are-still-good-eat-0154309/

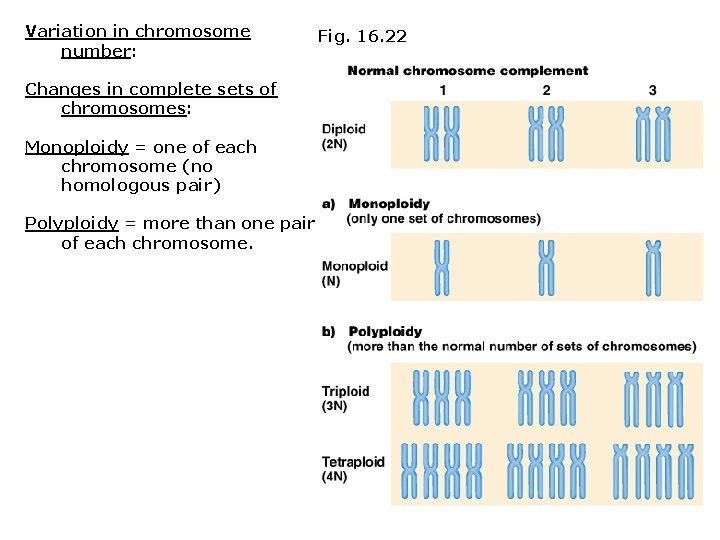
Variation in chromosome number: Changes in complete sets of chromosomes: Monoploidy = one of each chromosome (no homologous pair) Polyploidy = more than one pair of each chromosome. Fig. 16. 22

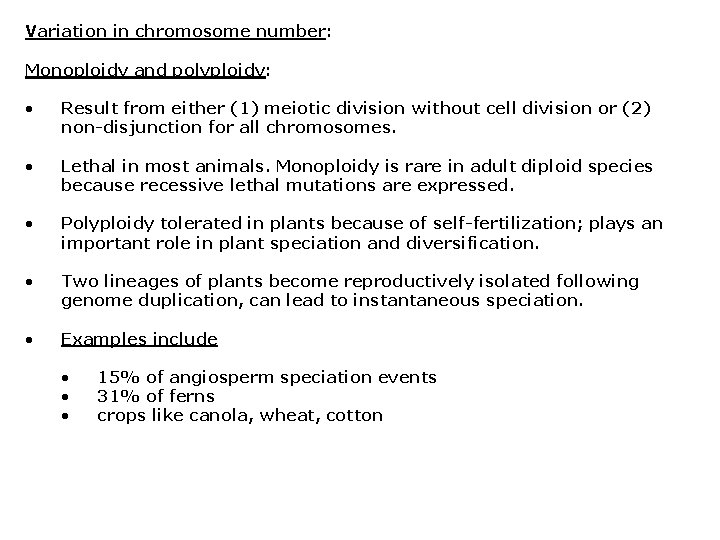
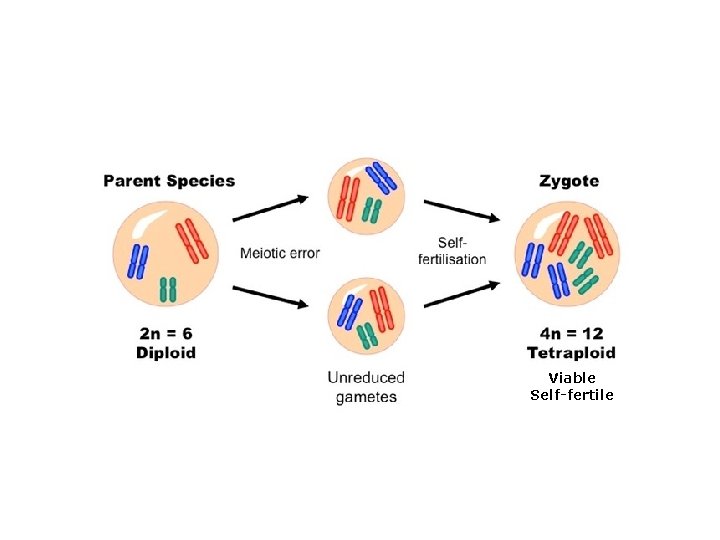
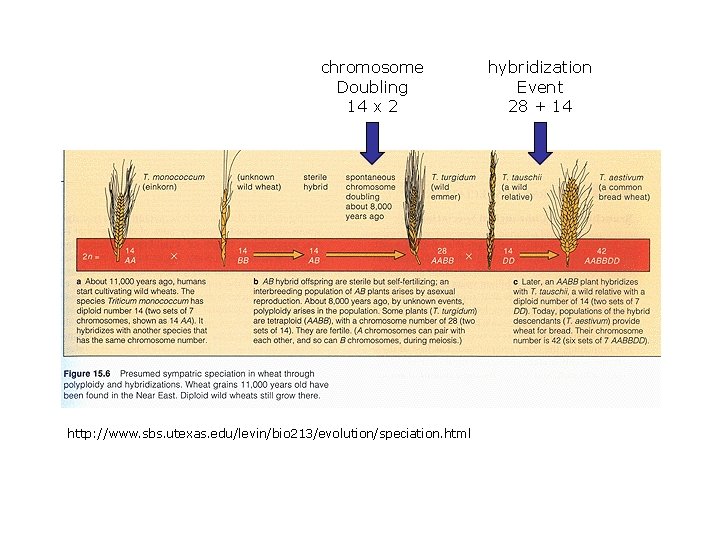
Variation in chromosome number: Monoploidy and polyploidy: • Result from either (1) meiotic division without cell division or (2) non-disjunction for all chromosomes. • Lethal in most animals. Monoploidy is rare in adult diploid species because recessive lethal mutations are expressed. • Polyploidy tolerated in plants because of self-fertilization; plays an important role in plant speciation and diversification. • Two lineages of plants become reproductively isolated following genome duplication, can lead to instantaneous speciation. • Examples include • • • 15% of angiosperm speciation events 31% of ferns crops like canola, wheat, cotton

Viable Self-fertile

chromosome Doubling 14 x 2 http: //www. sbs. utexas. edu/levin/bio 213/evolution/speciation. html hybridization Event 28 + 14

Odd-numbered polyploids have unpaired chromosomes and usually are sterile. Most seedless fruits we eat are triploid.