Chapter 15 GENE CONTROL I Prokaryotic Gene Control







- Slides: 34

Chapter 15 GENE CONTROL

I. Prokaryotic Gene Control � A. � � � Conserves Energy and Resources by 1. only transcribing genes when necessary a. don’t make m. RNA for tryptophan producing enzymes if tryptophan can be absorbed from environment 2. only producing proteins when needed a. don’t need lactose digesting enzymes if no lactose is present

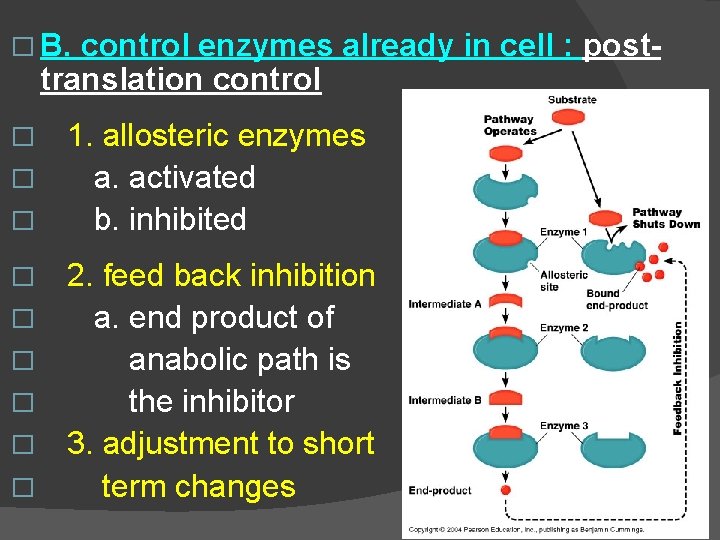
� B. control enzymes already in cell : posttranslation control 1. allosteric enzymes � a. activated � b. inhibited � � � � 2. feed back inhibition a. end product of anabolic path is the inhibitor 3. adjustment to short term changes

� C. control production of enzymes: transcription control � 1. control transcription of genes � a. repressors bind to operators and stop � transcription � b. enhancers bind to promotor to speed � transcription � 2. slower/longer lasting effects: more stable environment

� D. Negative control � 1. negative slows or stops function � 2. feed back inhibition (allosteric enzymes) � 3. repressors blocking transcription (gene control) � E. � � positive control 1. SPEEDS up production 2. just allowing production does not count!! 3. Enhancers bound to promoter 4. allosteric activators

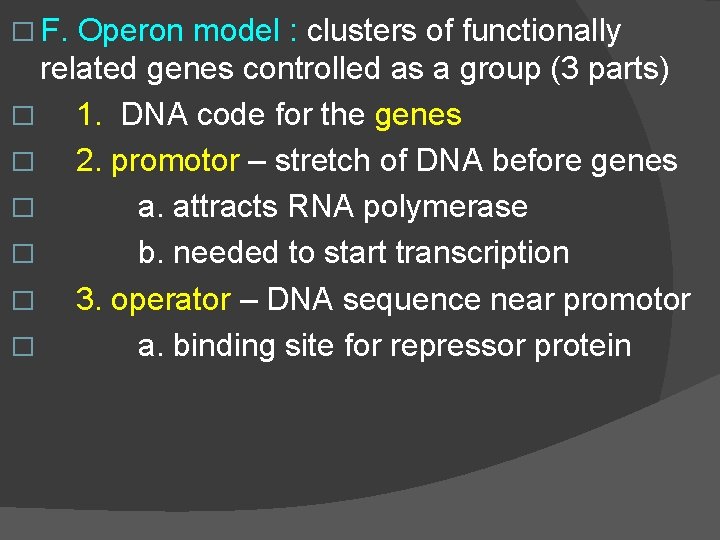
� F. Operon model : clusters of functionally related genes controlled as a group (3 parts) � 1. DNA code for the genes � 2. promotor – stretch of DNA before genes � a. attracts RNA polymerase � b. needed to start transcription � 3. operator – DNA sequence near promotor � a. binding site for repressor protein

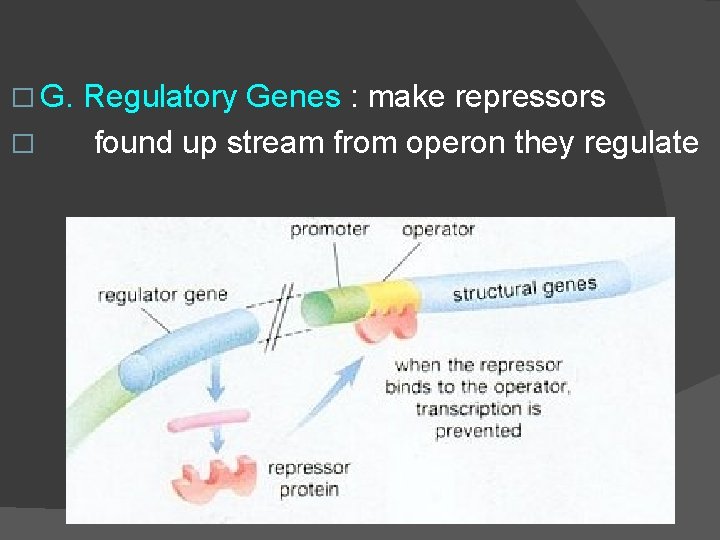
� G. � Regulatory Genes : make repressors found up stream from operon they regulate

� H. � � � trp Operon : trp = tryptophan amino acid 1. Repressible operon bcs it is normally active 2. genes make trp 3. low trp level in cell : operon active a. repressor is inactive b. promoter is open to RNA polymerase c. genes to make trp are copied

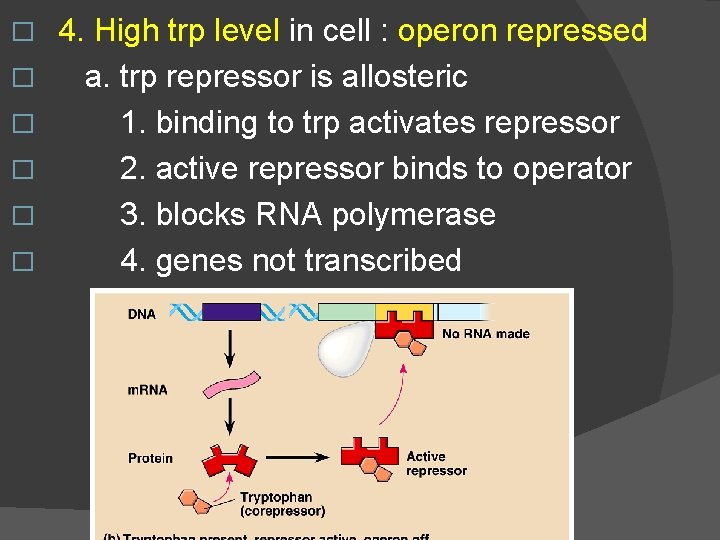
� � � 4. High trp level in cell : operon repressed a. trp repressor is allosteric 1. binding to trp activates repressor 2. active repressor binds to operator 3. blocks RNA polymerase 4. genes not transcribed

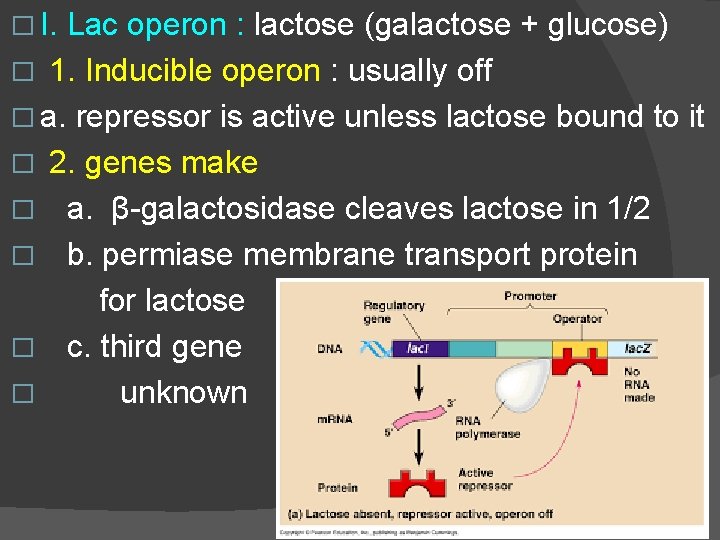
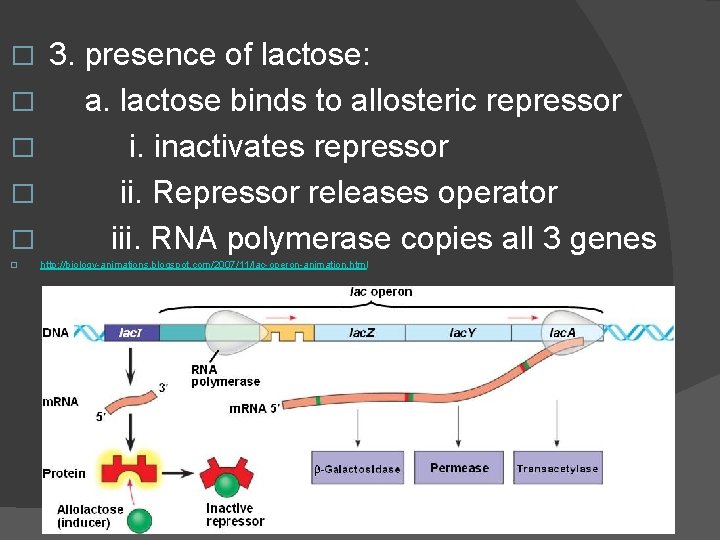
� I. Lac operon : lactose (galactose + glucose) � 1. Inducible operon : usually off � a. repressor is active unless lactose bound to it � 2. genes make � a. β-galactosidase cleaves lactose in 1/2 � b. permiase membrane transport protein for lactose � c. third gene � unknown

� � � 3. presence of lactose: a. lactose binds to allosteric repressor i. inactivates repressor ii. Repressor releases operator iii. RNA polymerase copies all 3 genes http: //biology-animations. blogspot. com/2007/11/lac-operon-animation. html

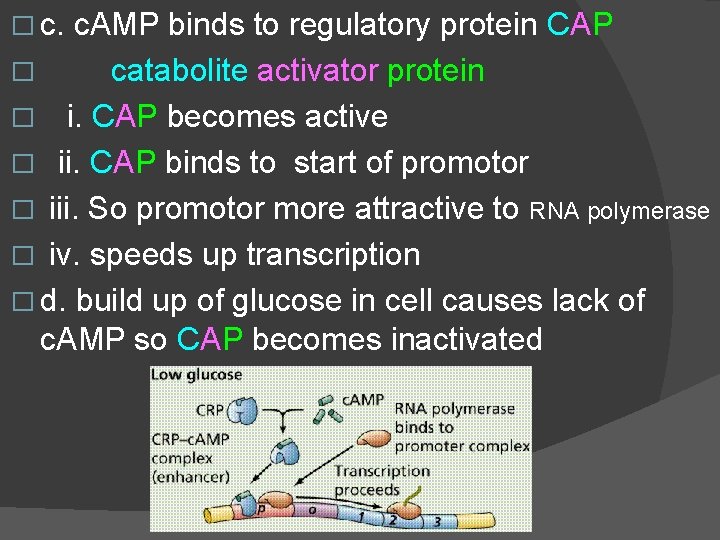
J. Positive control of lac operon � 1. lac operon is an inducible operon � = can be activated � 2. lac operon exhibits positive control � = can be speeded up � 3. If glucose is present E. coli prefer to use it � a. lack of glucose causes E. coli to speed up � use of lactose � b. lack of glucose causes build up of c. AMP � (cyclic AMP) = signal molecule � c. c. AMP signals speed up operon translation

� c. AMP binds to regulatory protein CAP � catabolite activator protein � i. CAP becomes active � ii. CAP binds to start of promotor � iii. So promotor more attractive to RNA polymerase � iv. speeds up transcription � d. build up of glucose in cell causes lack of c. AMP so CAP becomes inactivated

Prokaryotic Gene control review � Prokaryotic gene default setting is On/Off? � 2 main points at which Prokaryotes regulate gene expression? Transcription/translation/post-translation? � Transcription control � regulatory proteins bind to… � regulatory protein that exerts negative control = positive control = � Post-translation control: � regulatory molecule binds to… � regulatory molecule that exerts negative control = positive control =

Prokaryotic Gene control review � Prokaryotic gene default setting is On/Off? � 2 main points at which Prokaryotes regulate gene expression? Transcription/translation/post-translation? � Transcription control � regulatory proteins bind to… DNA � regulatory protein negative control = repressor � regulatory protein positive control = activator � Post-translation control: � regulatory molecule binds to…allosteric site � regulatory molecule negative control = inhibitor � regulatory molecule positive control = activator

� Repressible operons � a) repressor inactive w/o allosteric binding � b) normally on � c) usually anabolic � Inducible operons � a) repressor active unless bound � b) normally off � c) usually catabolic

II. Eukaryotic Gene Control � A. Gene expression regulated at many stages � 1. Transcription control � a. chromatin structure regulation � b. transcription initiation control � 2. Post-transcriptional control � a. RNA transcript processing � b. m. RNA degradation � c. Translation initiation � 3. Post-translational control a. allosteric P, b. P processing, c. P degradation

B. Chromatin structure control � 1. Heterochromatin – Chromatin that remains tightly compacted even in interphase � a. genes not transcribed � 2. acetylation – � a. acetyl group (-COCH 3) bonded to histone � b. loosens up chromatin winding � c. promotes transcription � 3. DNA methylation – � a. –CH 3 bonds to DNA blocking transcription � b. methylated regions passed on to daughter � cells


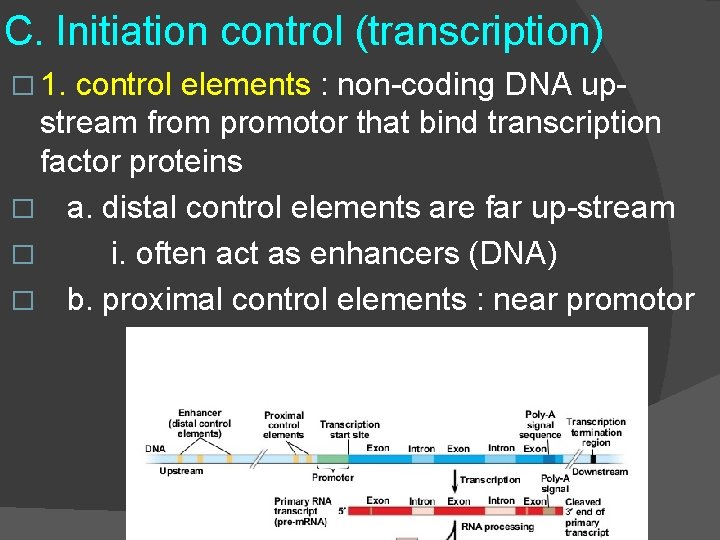
C. Initiation control (transcription) � 1. control elements : non-coding DNA upstream from promotor that bind transcription factor proteins � a. distal control elements are far up-stream � i. often act as enhancers (DNA) � b. proximal control elements : near promotor

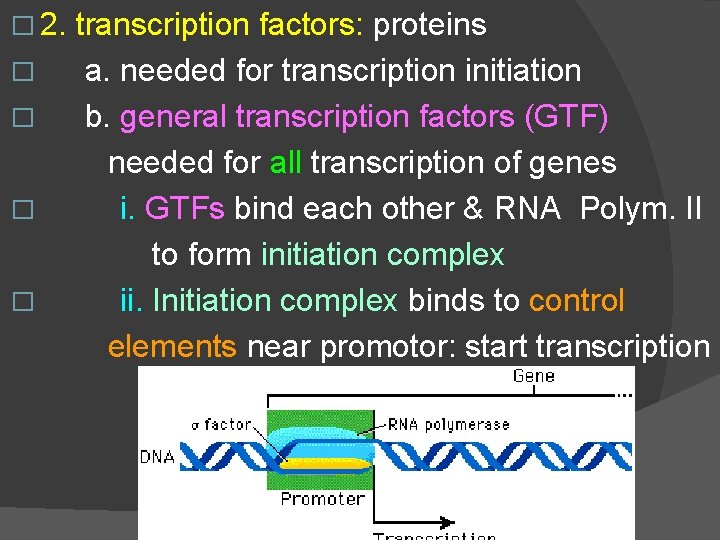
� 2. � � transcription factors: proteins a. needed for transcription initiation b. general transcription factors (GTF) needed for all transcription of genes i. GTFs bind each other & RNA Polym. II to form initiation complex ii. Initiation complex binds to control elements near promotor: start transcription

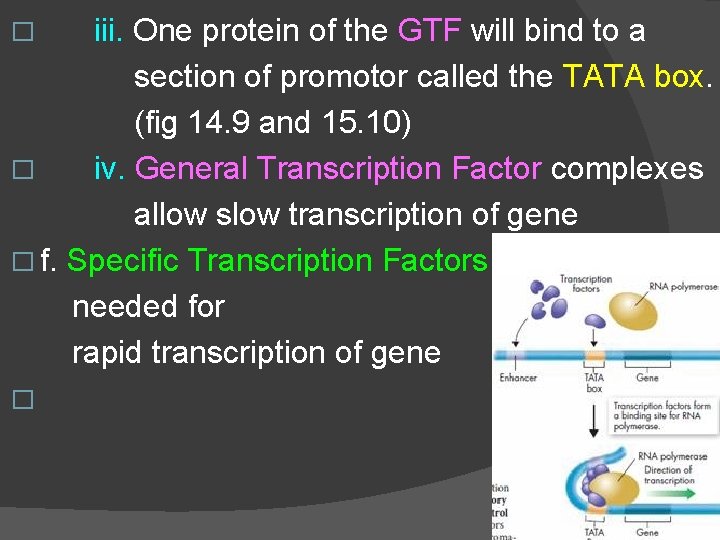
iii. One protein of the GTF will bind to a section of promotor called the TATA box. (fig 14. 9 and 15. 10) � iv. General Transcription Factor complexes allow slow transcription of gene � f. Specific Transcription Factors needed for rapid transcription of gene � �

� 3. Vocabulary in order to have a clue on 15. 2 � a. Things that are part of the DNA � i. control elements : binding site for transcription factors � ii. Enhancers : distal (far) control elements, can be activated or repressed by transcription factor proteins � iii. TATA box : section of the promoter’s code � iv. promoter : just upstream from start of gene, where RNA polymerase binds to start transcription

b. Things that are proteins � i. Transcription factor : regulatory protein binds control elements � a. general transcription factors allow transcription � b. activators speed transcription � c. repressors slow transcription � ii. Mediator proteins : form link between regulatory proteins and DNA �

� 4. Distal control elements = enhancers � a. may be up or down stream � b. each gene can have many enhancers � i. each active under different conditions � ii. Or active in different cell types � iii. Each enhancer works with only one gene � c. transcription factors called activator proteins bind to enhancer control elements � i. fold DNA so that the activator protein/enhancer complex binds to initiation complex to speed up transcription


� d. repressor transcription factors interfere with the activator transcription factors to slow transcription � i. by binding to distal control elements and keeping activators out � ii. By binding to activator proteins

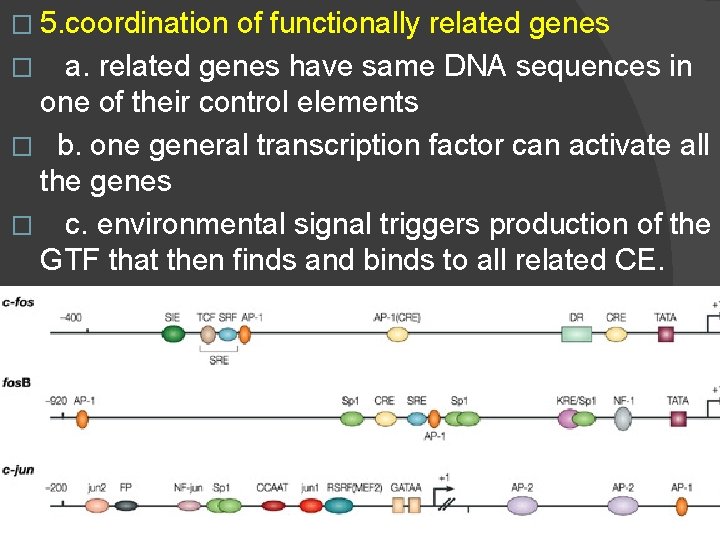
� 5. coordination of functionally related genes � a. related genes have same DNA sequences in one of their control elements � b. one general transcription factor can activate all the genes � c. environmental signal triggers production of the GTF that then finds and binds to all related CE.

D. Post-transcription Control � 1. RNA processing � a. alternative splicing � b. poly A tail length � c. cap designation � 2. m. RNA degradation � a. specialized RNAs can degrade m. RNA � 3. Translation initiation control � a. proteins bond to m. RNA prevent initiation � b. egg m. RNA � c. global control : lack of initiation factor (egg)

E. Post Translation Control : Protein Processing/degradation � 1. Allosteric control or activation by phosphate � 2. protein processing a. inactive form cut to activate (pro-insulin) b. glycoproteins, lipoproteins � � 3. selective degradation � a. ubiquitin = protein attached to proteins tags them for destruction �

F. Non-coding RNA (nc. RNA) 1)Don’t code for proteins � � � � a) micro. RNAs (mi. RNAs) i. complexes w/ proteins ii. binds to complementary m. RNA iii stops translation or trigger degradation b) small interfering RNA (si. RNA) i. turn off gene expression ii. Used in knock-out experiments c) nc. RNA affect heterochromatin formation

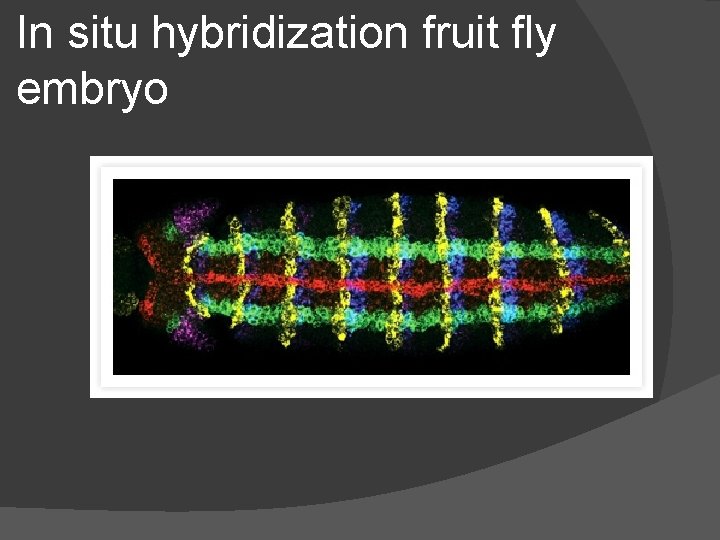
G. Monitoring gene expression � 1) To see which cells are using a gene do… � in situ hybridization (in situ = in place) � a. need to detect m. RNA made from that gene � b. hybridization = bonding complementary bases to a template � c. the m. RNA from the gene of interest = template � d. complementary strand = probe (DNA or RNA) � c. fluorescent dye added to probe � d. probe added to solution around embryo � b. probe hybridizes to target m. RNA & concentrates in cells that are using the gene of interest

In situ hybridization fruit fly embryo

� 2) reverse transcriptase – PCR (RT-PCR) � a. used to see how much m. RNA is present � b. isolate m. RNA and use reverse transcriptase to make c. DNA (complimentary DNA) � c. use PCR and electrophoresis to determine how much m. RNA was present � 3) RNA sequencing : sequence c. DNA