Chapter 13 Molecular Basis of Inheritance The Search
Chapter 13: Molecular Basis of Inheritance

The Search for the Genetic Material • 1928 = Frederick Griffith and Streptococcus pneumoniae • 1940 s = Protein or DNA ? ? – The case for proteins seemed stronger • 1944 = Oswald Avery, Maclyn Mc. Carty and Colin Mac. Leod working with E. coli and T 2 • 1953 = James Watson and Francis Crick shook the scientific world with their elegant DNA model

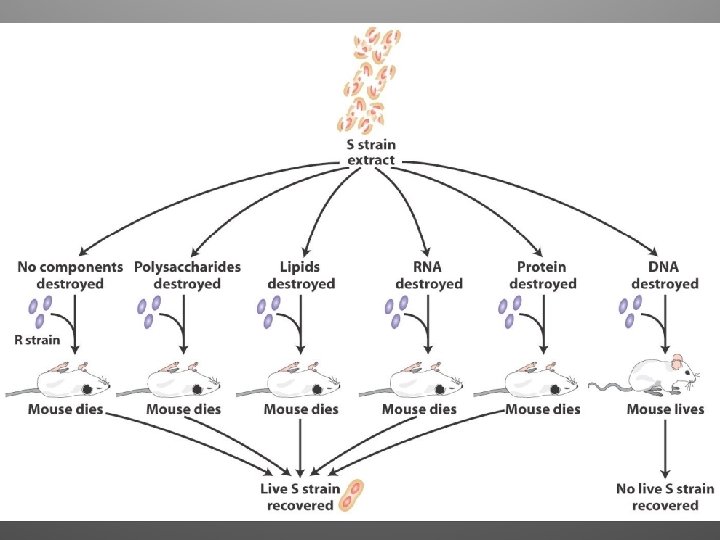
Frederick Griffith, 1928 • Attempting to develop a vaccine against pneumonia • Two strains: – One pathogenic (S strain, capsule) – One non pathogenic (R strain) • S train was killed by heating, and mixed the dead S strain with live R strain • Transformation occurred!!

Streptococcus pneumoniae
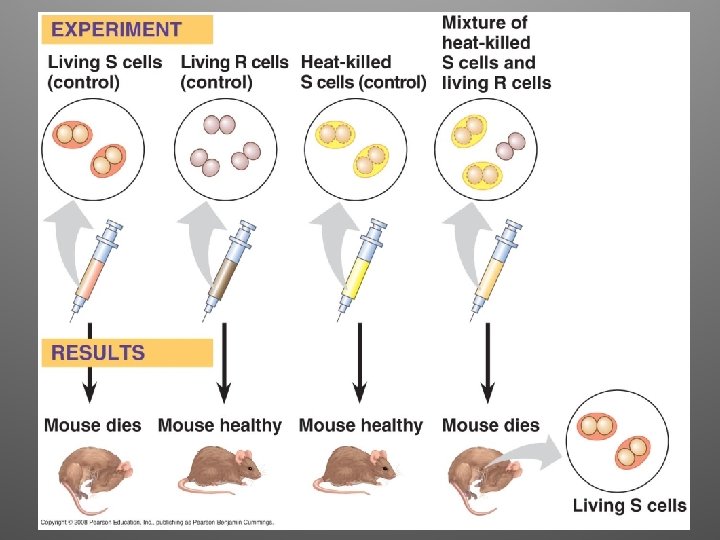
Observations • Clearly, some chemical component of the dead pathogenic cells caused this heritable change • Griffith called the phenomenon transformation (assimilation of external DNA that causes change in genotype and phenotype)

Avery, Mc. Carty and Mac. Leod 1944 • Studied DNA, RNA and protein for 14 years • They worked with Streptococcus pneumoniae • In 1944 Avery, Mac. Leod and Mc. Carty announced that the transforming agent was DNA • Their discovery was greeted with interest but considerable skepticism

Oswald Avery Colin Mac. Leod Maclyn Mc. Carty

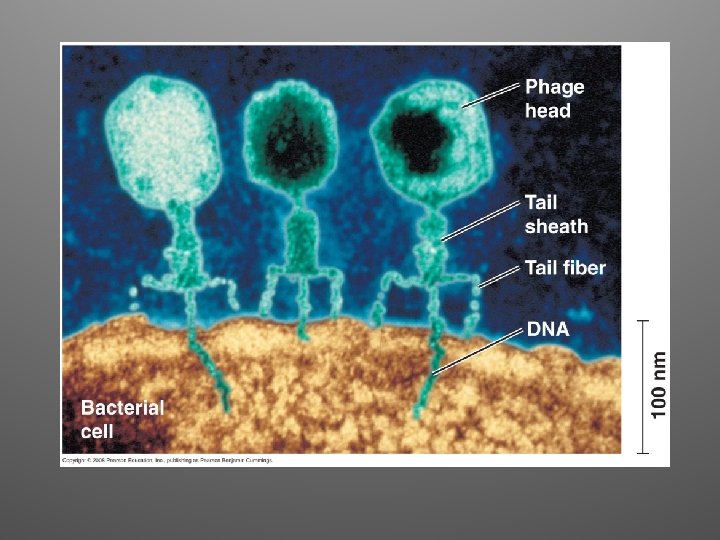
Alfred Hershey and Martha Chase, 1952 • They worked with bacteriophages (phages) and Escherichia coli (intestines of mammals) • T 2 sequesters E. coli’s cell machinery and makes it produce many copies of itself

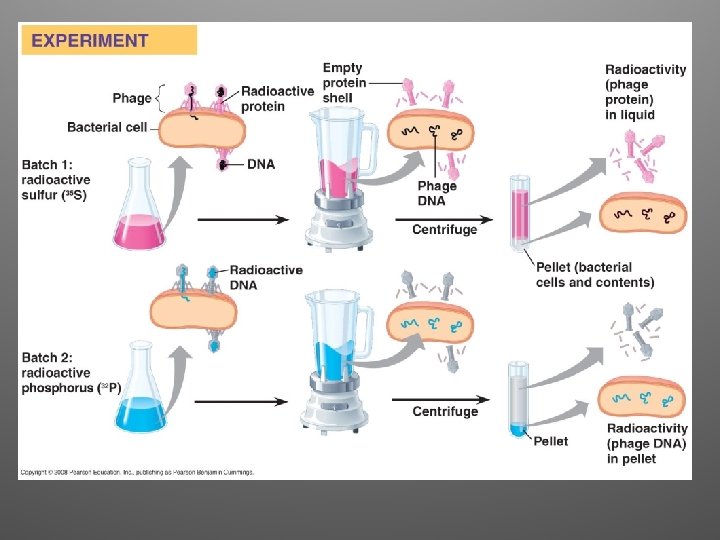
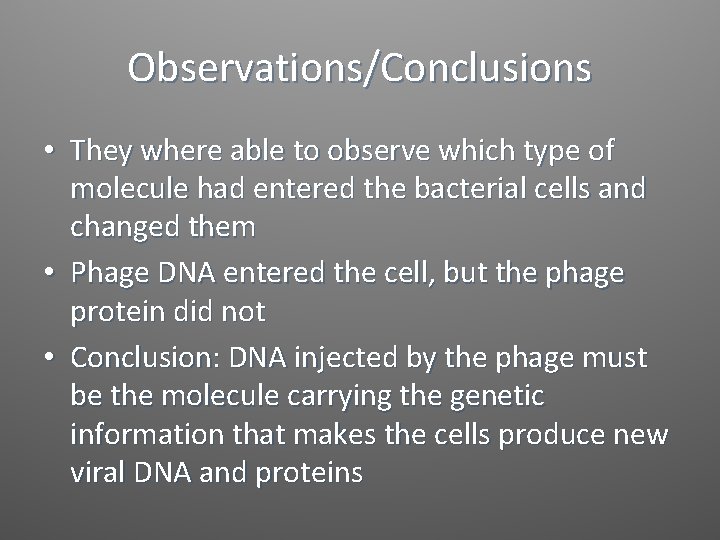
Observations/Conclusions • They where able to observe which type of molecule had entered the bacterial cells and changed them • Phage DNA entered the cell, but the phage protein did not • Conclusion: DNA injected by the phage must be the molecule carrying the genetic information that makes the cells produce new viral DNA and proteins

Erwin Chargaff, 1950
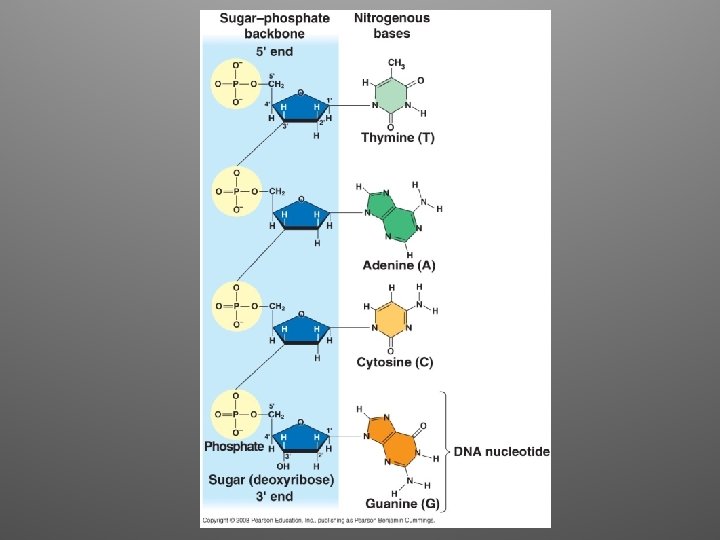
Chargaff • It was already know that DNA is a polymer of nucleotides • Analyzed the base composition of DNA from a number of different organisms • He noticed a peculiar regularity in the ratios of nucleotide bases within a single species
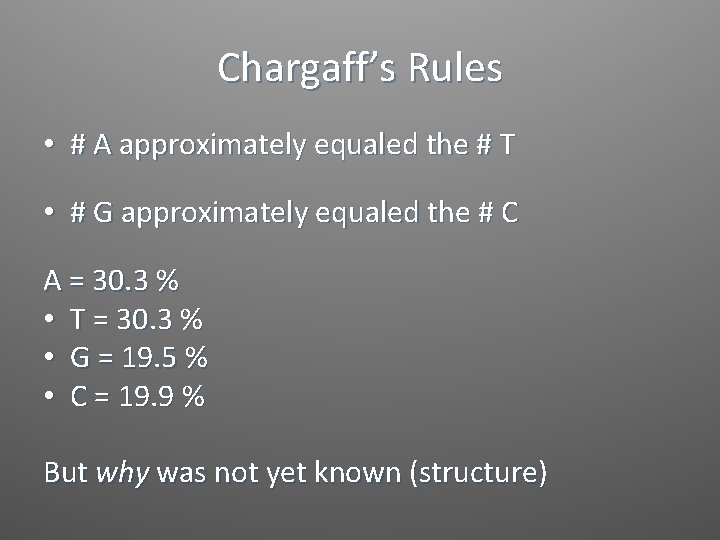
Chargaff’s Rules • # A approximately equaled the # T • # G approximately equaled the # C A = 30. 3 % • T = 30. 3 % • G = 19. 5 % • C = 19. 9 % But why was not yet known (structure)

Building a DNA model (1950 s) • Linus Pauling proposed a triple helix model (California Institute of Technology) • Maurice Wilkins and Rosalind Franklin (Kings College, London)

The story… • James Watson journeyed to Cambridge, where Francis Crick was studying protein structure using X-ray crystallography • Watson visited Maurice Wilkins Lab and saw the diffractions images produced by Franklin
X-ray Chrystallography • Produces an X-ray diffraction image • The crystal is bombarded with X-rays • The rays are deflected as the pass through the aligned fibers of purified DNA • Mathematical equations are used to translate the patterns into information on the 3 D structure of the molecule

Watson and Crick • Built models of a double helix that would conform to the X-ray measurements and to what was known about the chemistry of DNA • They read an unpublished annual report summarizing Franklin’s work where she had concluded that the sugar-phosphate backbones were on the outside of the double helix
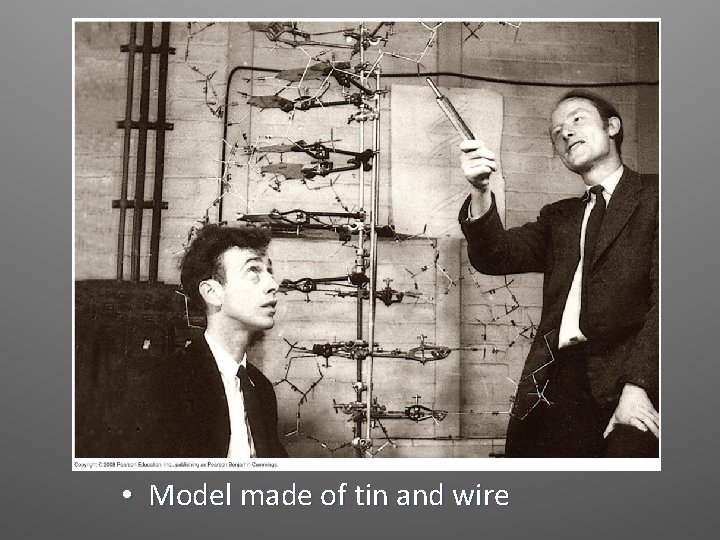
• Model made of tin and wire

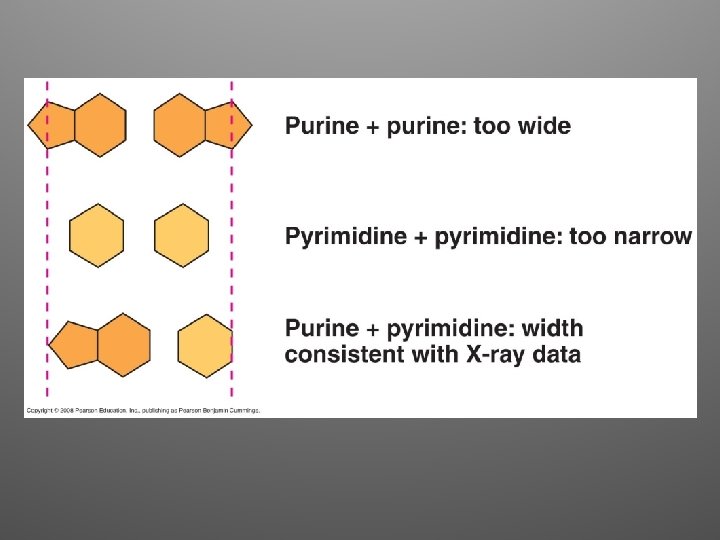
pyrimidine purine
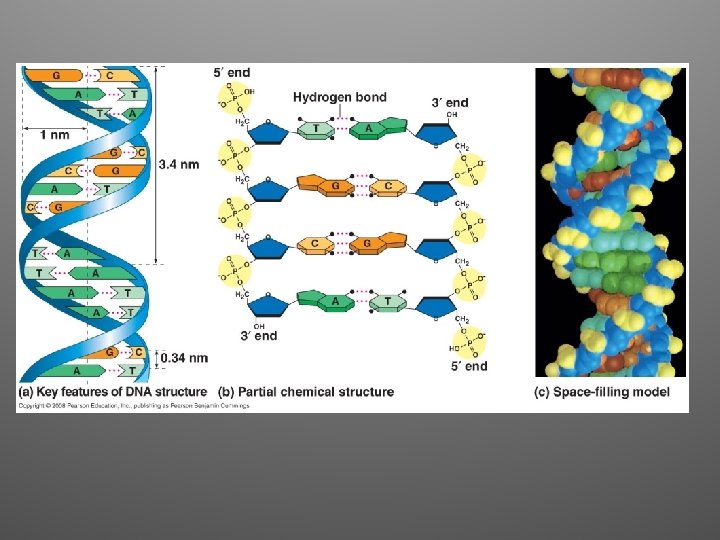

Great idea!!! • The arrangement was appealing because it put the relatively hydrophobic nitrogenous bases towards the interior • Sugar-phosphate backbones are antiparallel • Nitrogenous bases are paired in specific combinations (maintain uniform diameter)

April 1953 • Watson and Crick surprised the scientific world with a succinct, one-page paper in Nature • The beauty of the model is that is suggested a mechanism for replication
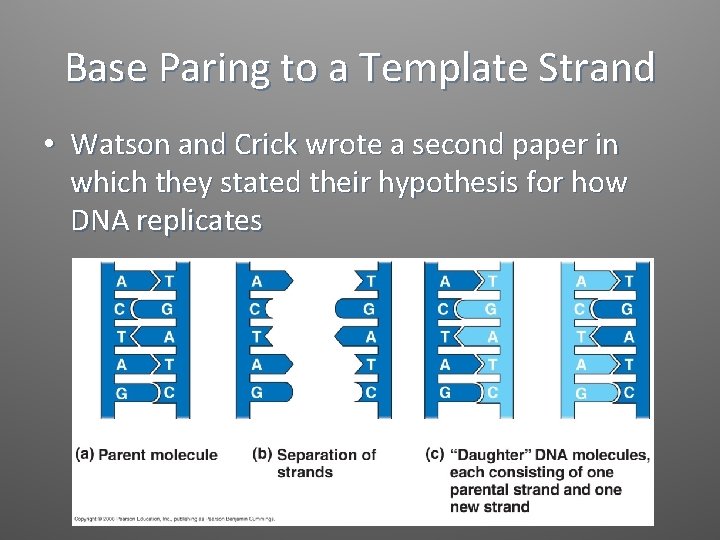
Base Paring to a Template Strand • Watson and Crick wrote a second paper in which they stated their hypothesis for how DNA replicates
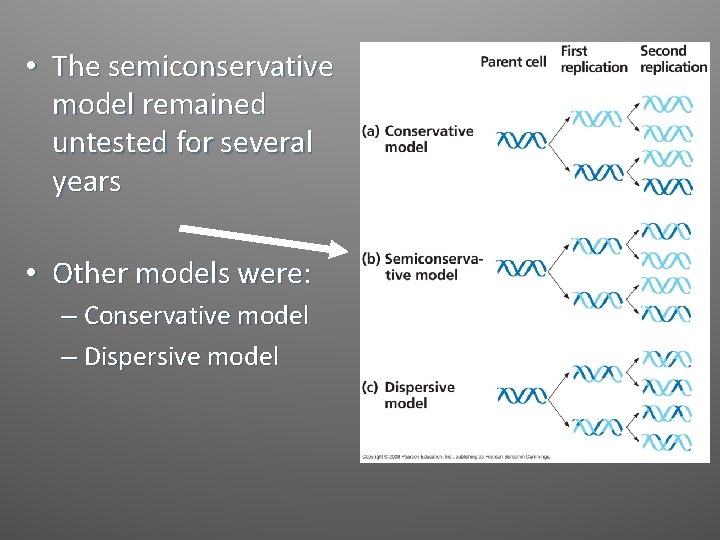
• The semiconservative model remained untested for several years • Other models were: – Conservative model – Dispersive model

1962: Nobel Prize awarded • Watson and Crick • Also to Maurice Wilkins • But not given to Rosalind Franklin! – She sadly died of cancer in 1958
What is the genetic material for life? A. B. C. D. RNA DNA Proteins Lipids

Matthew Meselson and Franklin Stahl

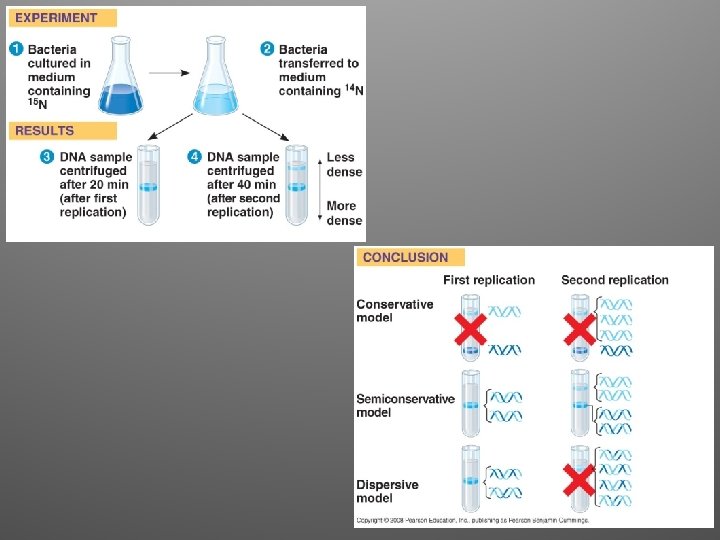
Matthew Meselson and Franklin Stahl • In 1958 they published a clever experiment that distinguished between the three models • Their results supported the semiconservative model of replication
DNA replication: a closer look • E. coli has a singular, circular chromosome of about 4. 6 million nucleotide pairs • It can divide to form two genetically identical daughter cells in less than an hour • More than a dozen enzymes and other proteins participate in DNA replication
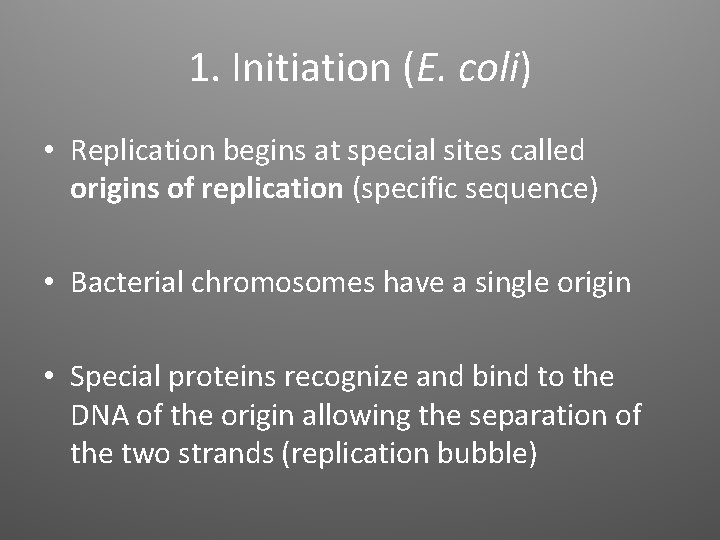
1. Initiation (E. coli) • Replication begins at special sites called origins of replication (specific sequence) • Bacterial chromosomes have a single origin • Special proteins recognize and bind to the DNA of the origin allowing the separation of the two strands (replication bubble)
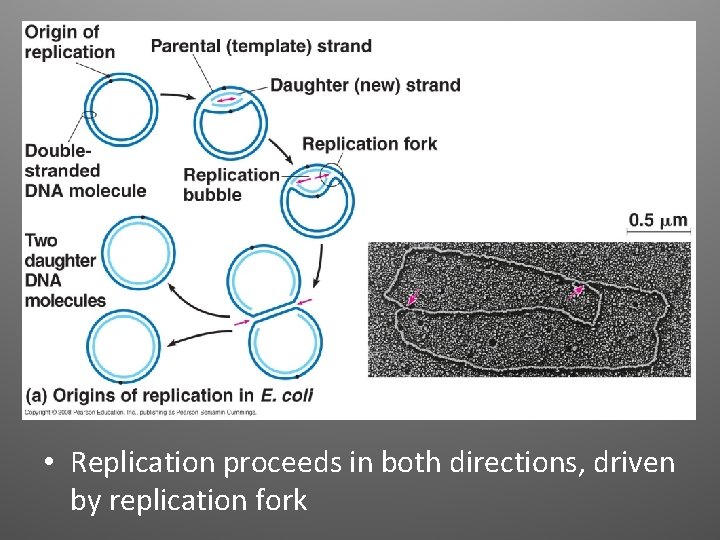
• Replication proceeds in both directions, driven by replication fork
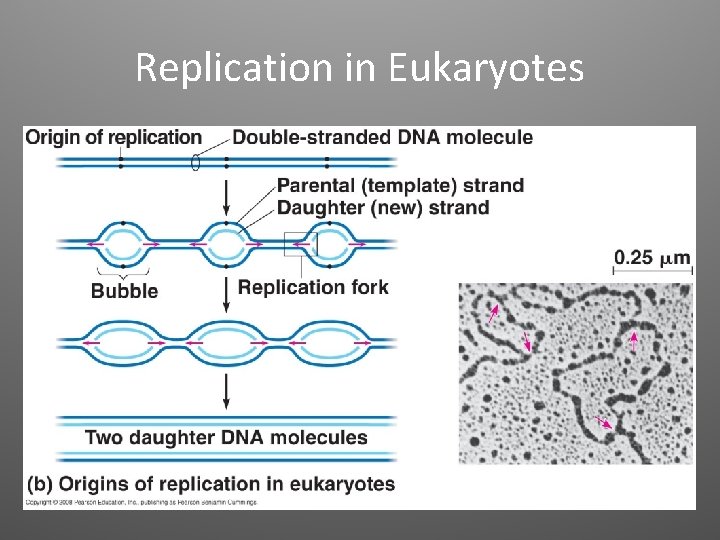
Replication in Eukaryotes
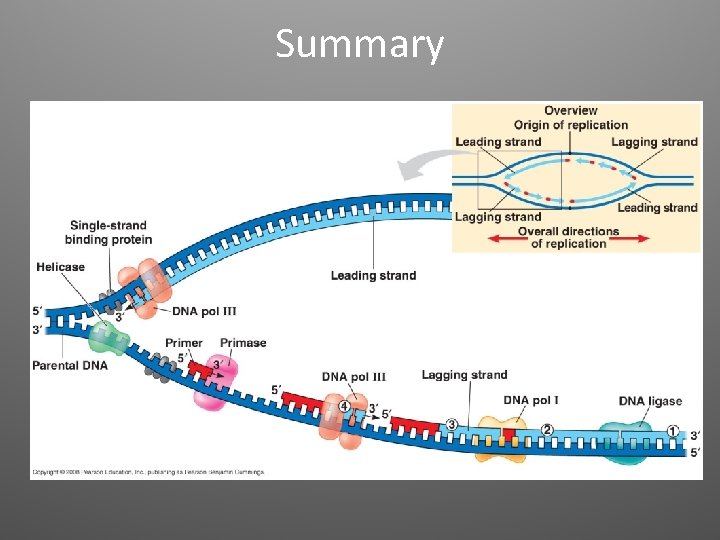
Other proteins • Helicases: untwist the double helix at the replication forks • Single-strand binding proteins: bind to the unpaired DNA strands and stabilize them • Topoisomerase: relives super-coiling • Primase: allow initiation of synthesis by adding a short stretch of RNA (primer, 5 -10 nt)
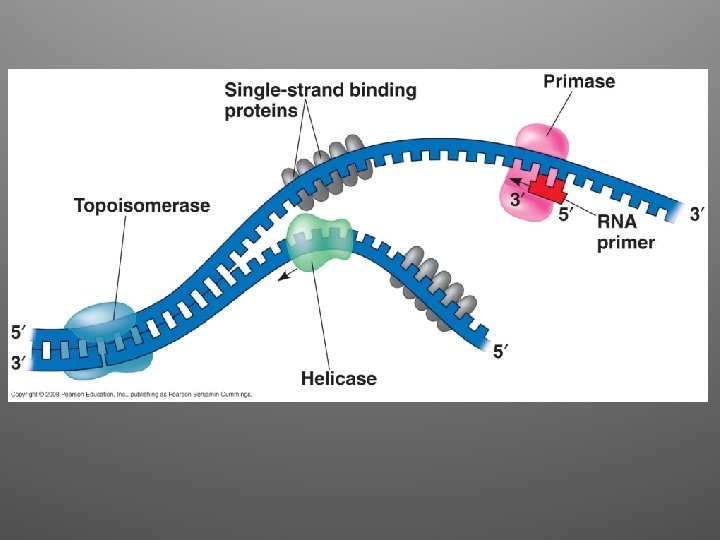
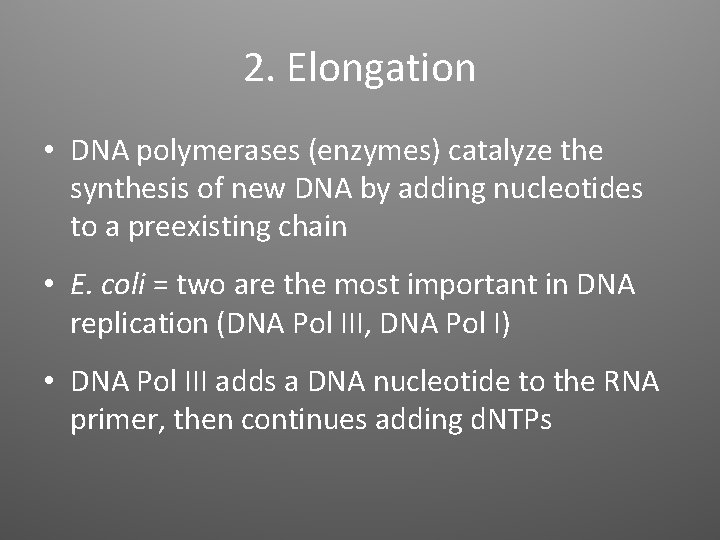
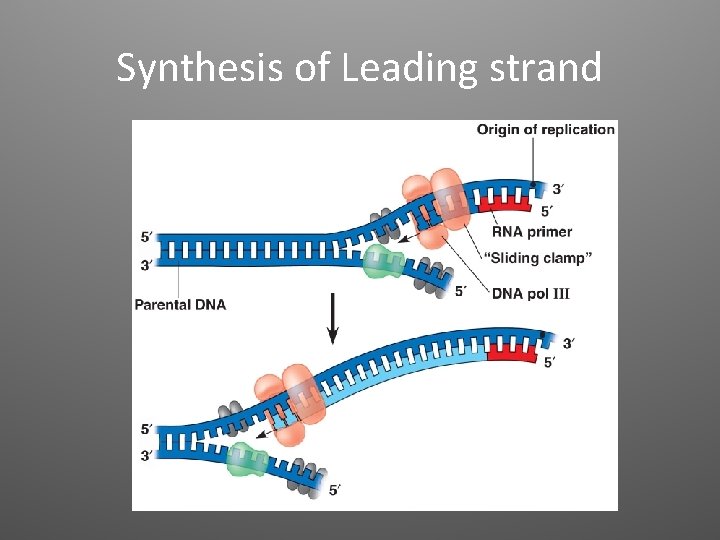
2. Elongation • DNA polymerases (enzymes) catalyze the synthesis of new DNA by adding nucleotides to a preexisting chain • E. coli = two are the most important in DNA replication (DNA Pol III, DNA Pol I) • DNA Pol III adds a DNA nucleotide to the RNA primer, then continues adding d. NTPs
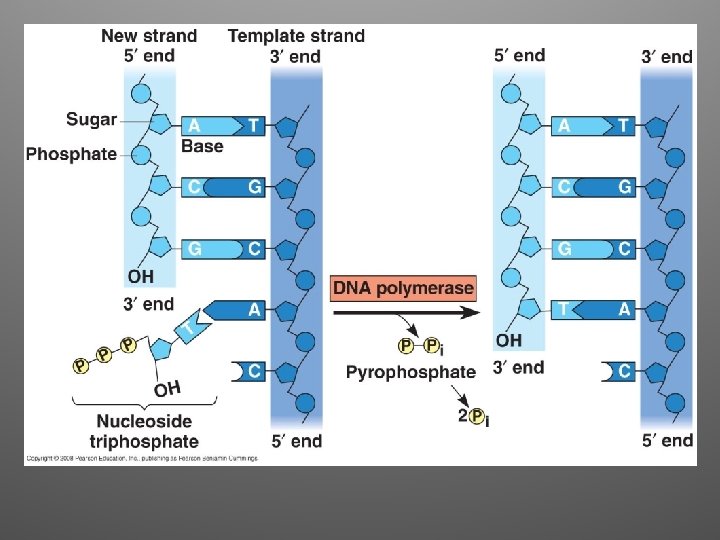
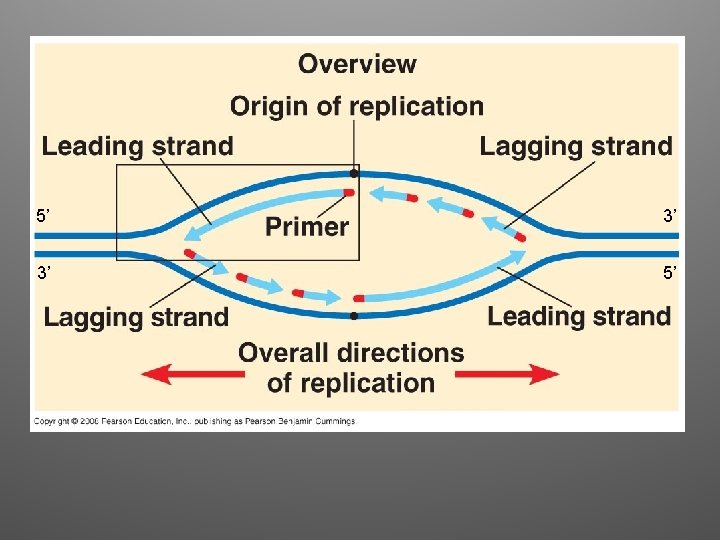
Antiparallel Elongation • The two strands of the double helix are antiparallel • DNA Pol can only add nucleotides to a free 3’ end (5’ to 3’ direction) • Elongation: – Leading strand: continuous elongation, only one RNA primer – Lagging strand: discontiuous elongation, many RNA primers
Synthesis of Leading strand

Synthesis of Lagging strand
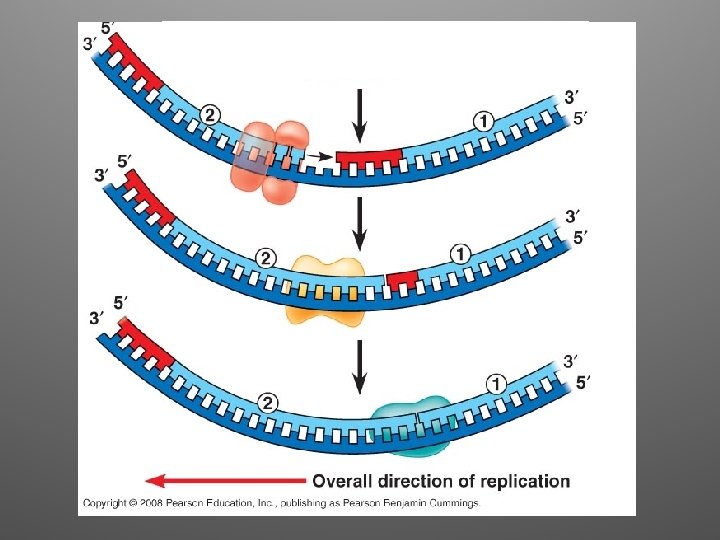

Lagging Strand • DNA Pol moves away from the replication fork • The segments observed on the lagging strand are called Okazaki Fragments (1000 -2000 nt) • DNA Pol I removes RNA nucleotides • DNA ligase joins the final nucleotide of the replaced segment
Summary
DNA Replication Complex • It is convenient to represent DNA polymerase molecules as little trains, but: – Various proteins actually form a single large complex – DNA replication complex does not move: rather, DNA moves through the complex (in eukaryotes, they may be anchored to the nuclear matrix) – Lagging strand is looped back through complex

Proofreading and Repair • There is an error rate of about 1/100, 000 nucleotides • During DNA replication, DNA polymerases proofread each nucleotide against its template • Removes incorrect nucleotides and resumes synthesis
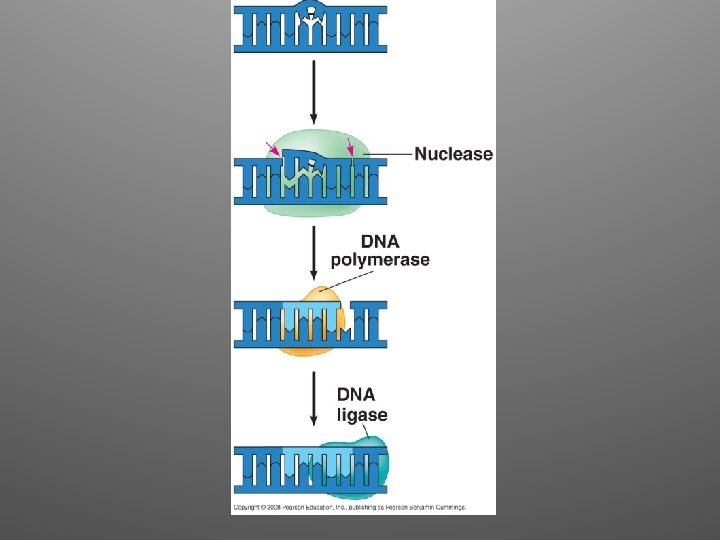
Repair Mechanisms • Mismatch Repair: – Enzymes remove and replace the incorrectly paired nucletide • Nucleotide Excision Repair: – A nuclease cuts out the damaged strand the nucleotides are replaced (Ex: thymine dimers)
Replicating the Ends of DNA Molecules • DNA Pol can only add nucleotides to a 3’ end • This represents a problem for linear chromosomes (eukaryotes) • Once the last primer is removed, it cannot be replaced with DNA • After repeated rounds of replication, shorter, uneven DNA molecules remain
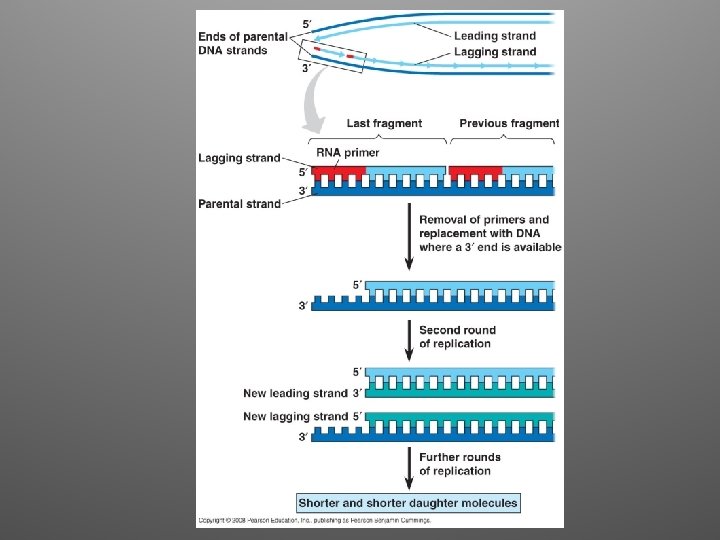
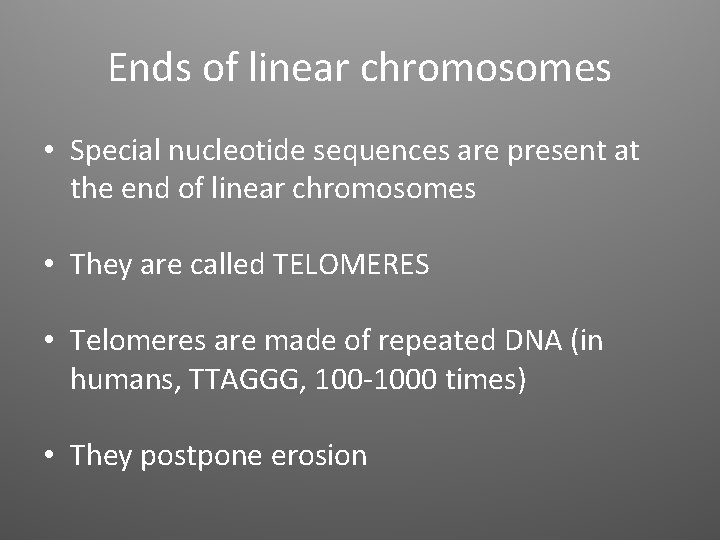
Ends of linear chromosomes • Special nucleotide sequences are present at the end of linear chromosomes • They are called TELOMERES • Telomeres are made of repeated DNA (in humans, TTAGGG, 100 -1000 times) • They postpone erosion
What about germ cells? • An enzyme called telomerase catalyzes the lengthening of telomeres in eukaryotic germ cells • They compensate for shortening that occurs during DNA replication • Inactive in most human somatic cells • Active in cancer cells
Chromosome Structure • Bacterial chromosomes: – circular, – double stranded – associate with a small amount of protein • Eukaryotic chromosomes: – Linear DNA – Associate with large amount of protein

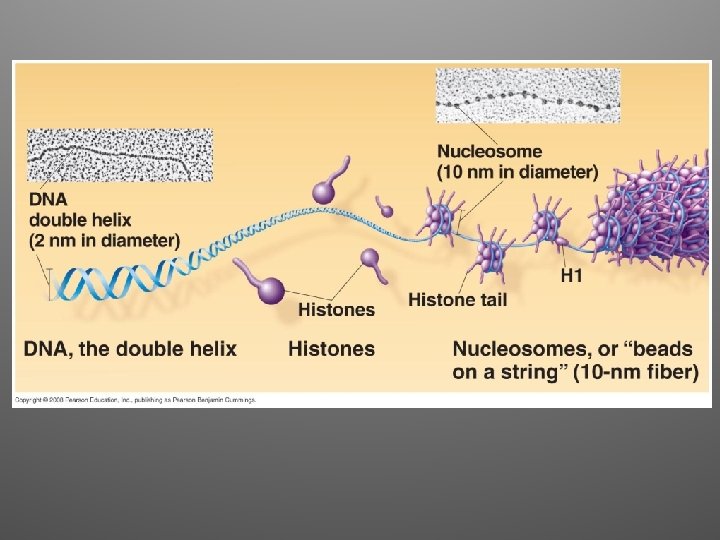
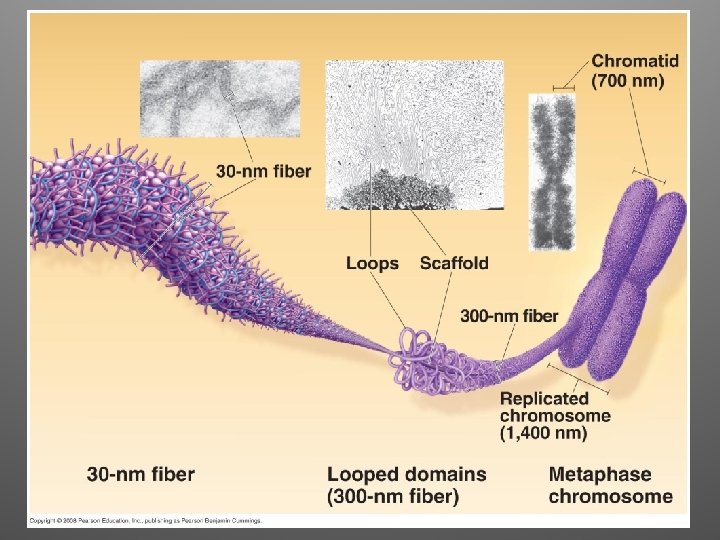
Eukaryotic Chromosomes • DNA + protein = chromatin • Multilevel DNA packing system: – DNA – 10 nm fiber – 30 nm fiber – Looped domains – Metaphase chromosome
Eukaryotic Chromosomes • Chromatin of each chromosome occupies a specific restricted area within the interphase nucleus (they do not become entangled) • Condensed state: – Heterochromatin: highly condensed – Euchromatin: more dispersed. DNA is accessible for expression machinery
- Slides: 65