Chapter 12 DNA Structure and Replication Identifying the







- Slides: 45

Chapter 12: DNA Structure and Replication

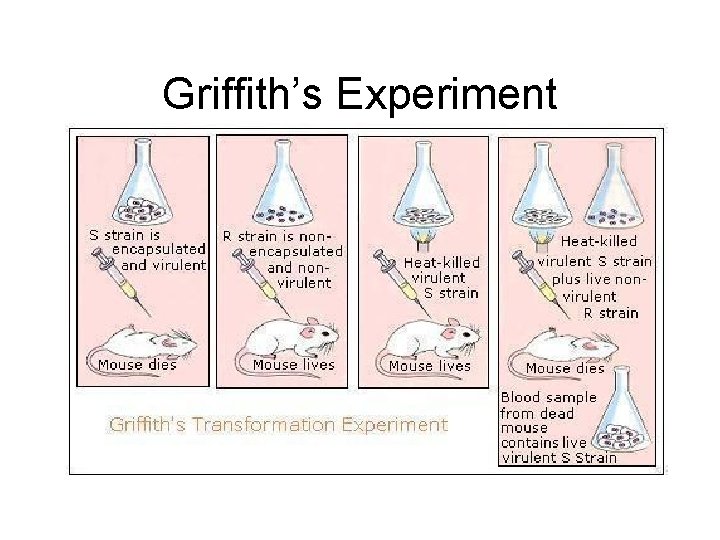
Identifying the substances of genes • • • Fredrick Griffith (1928) Tried to figure out what was getting people sick. Conducted an experiment with mice and discovered transformation. – When one type of bacteria (healthy) is changed into another (diseased) – Concluded that the transforming factor had to be a gene.

Griffith’s Experiment

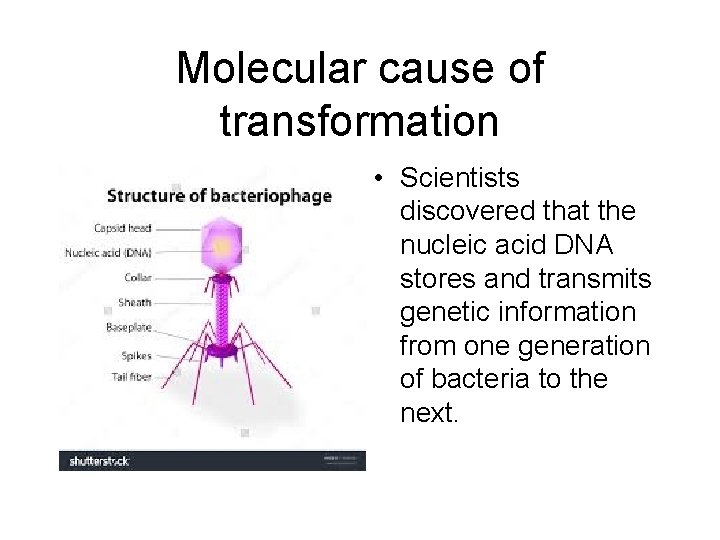
Molecular cause of transformation • Scientists discovered that the nucleic acid DNA stores and transmits genetic information from one generation of bacteria to the next.

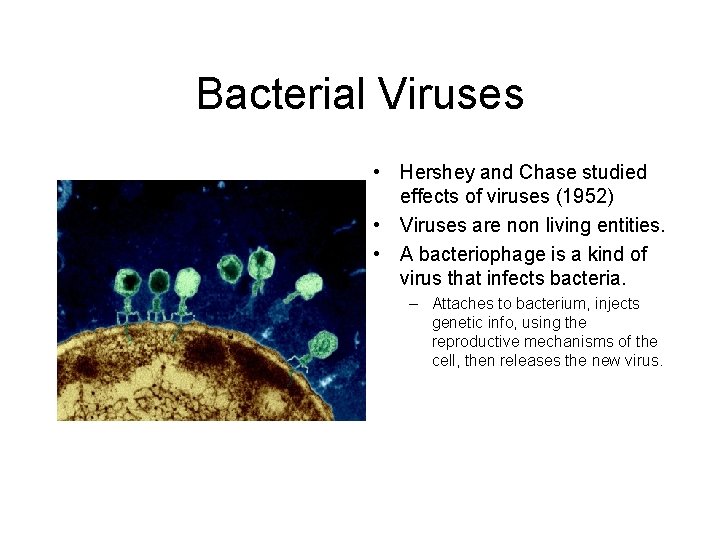
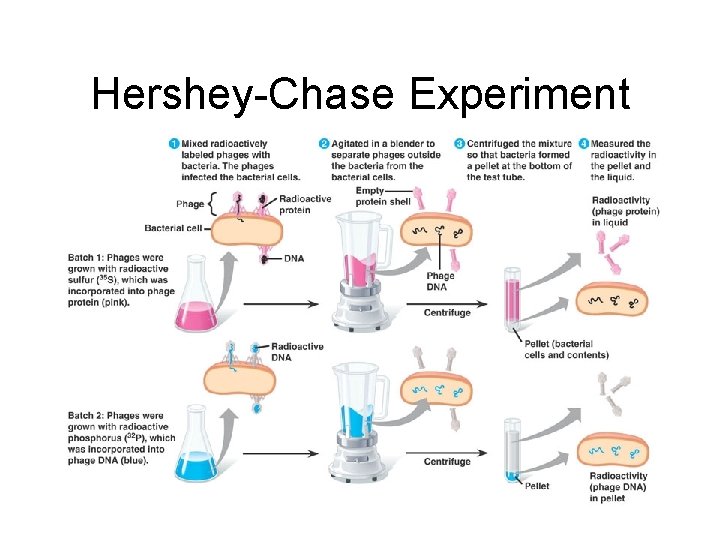
Bacterial Viruses • Hershey and Chase studied effects of viruses (1952) • Viruses are non living entities. • A bacteriophage is a kind of virus that infects bacteria. – Attaches to bacterium, injects genetic info, using the reproductive mechanisms of the cell, then releases the new virus.

Hershey-Chase Experiment

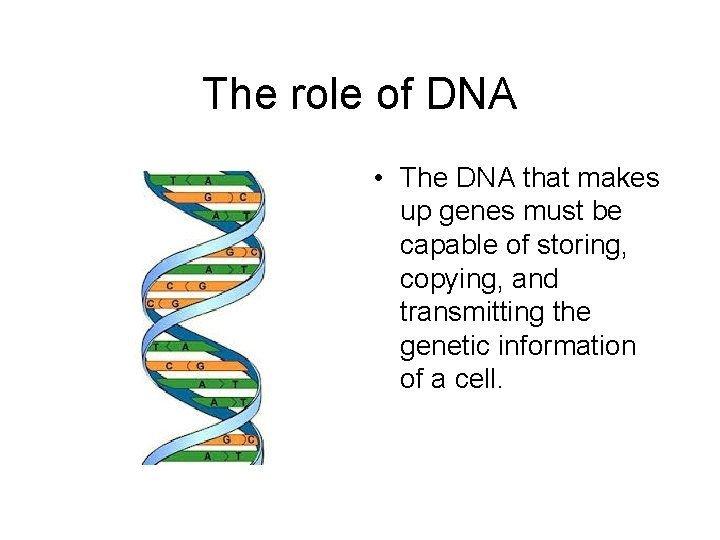
The role of DNA • The DNA that makes up genes must be capable of storing, copying, and transmitting the genetic information of a cell.

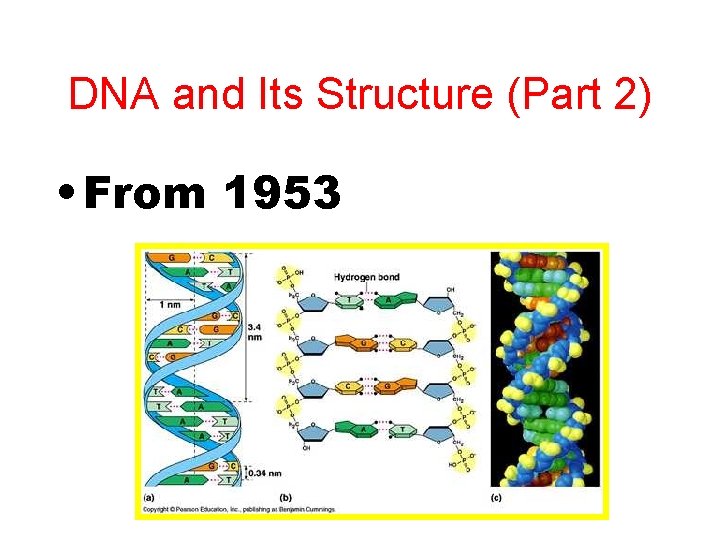
DNA and Its Structure (Part 2) • From 1953

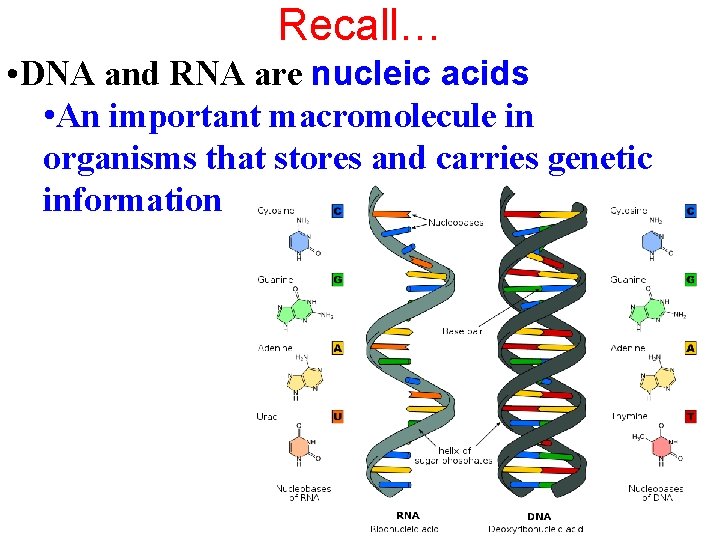
Recall… • DNA and RNA are nucleic acids • An important macromolecule in organisms that stores and carries genetic information

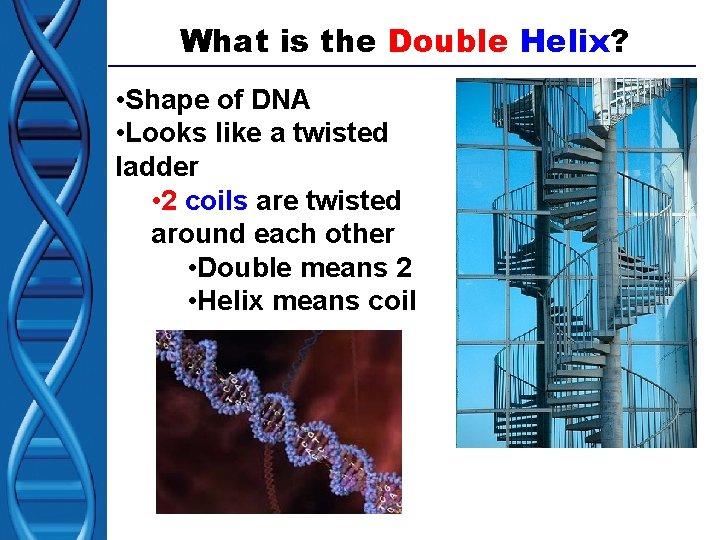
What is the Double Helix? • Shape of DNA • Looks like a twisted ladder • 2 coils are twisted around each other • Double means 2 • Helix means coil

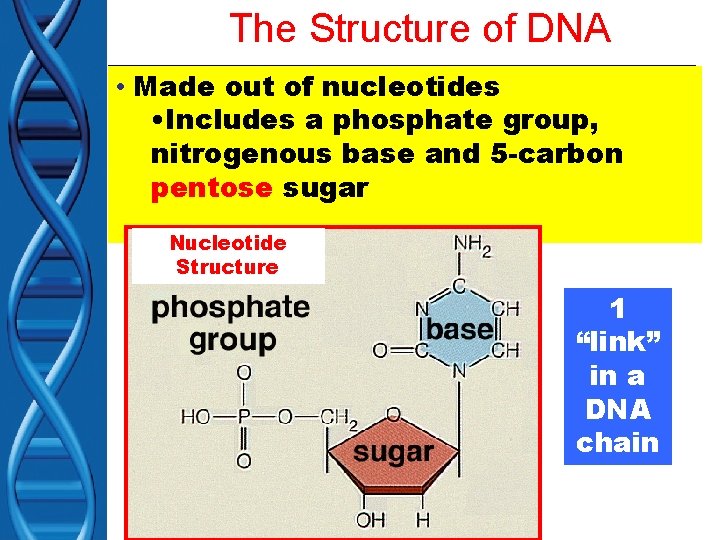
The Structure of DNA • Made out of nucleotides • Includes a phosphate group, nitrogenous base and 5 -carbon pentose sugar Nucleotide Structure 1 “link” in a DNA chain

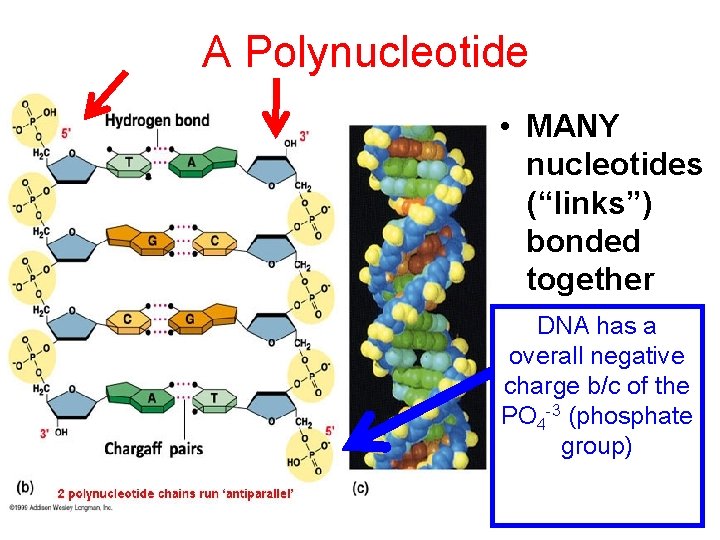
A Polynucleotide • MANY nucleotides (“links”) bonded together DNA has a overall negative charge b/c of the PO 4 -3 (phosphate group)

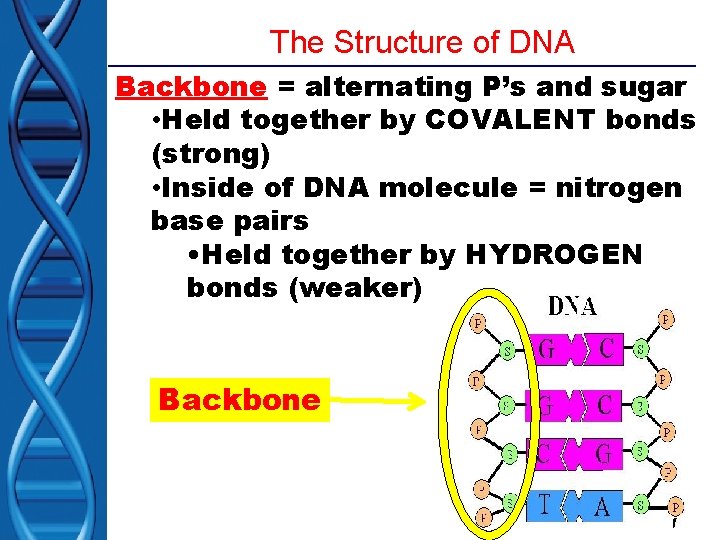
The Structure of DNA Backbone = alternating P’s and sugar • Held together by COVALENT bonds (strong) • Inside of DNA molecule = nitrogen base pairs • Held together by HYDROGEN bonds (weaker) Backbone

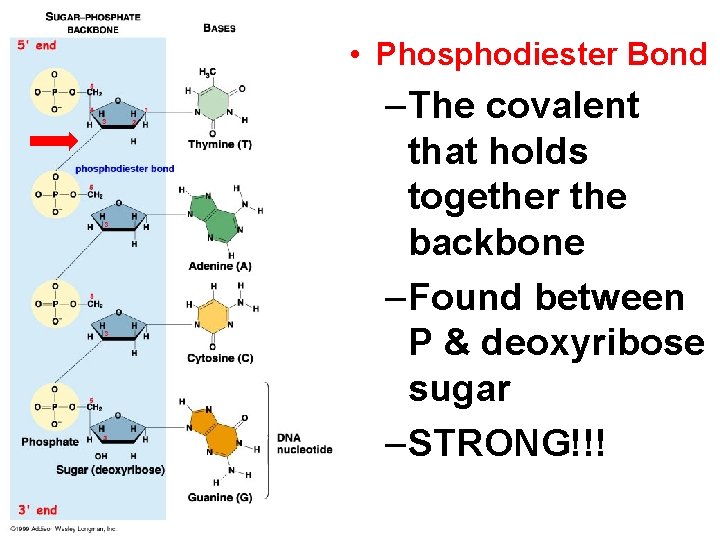
• Phosphodiester Bond –The covalent that holds together the backbone –Found between P & deoxyribose sugar –STRONG!!!

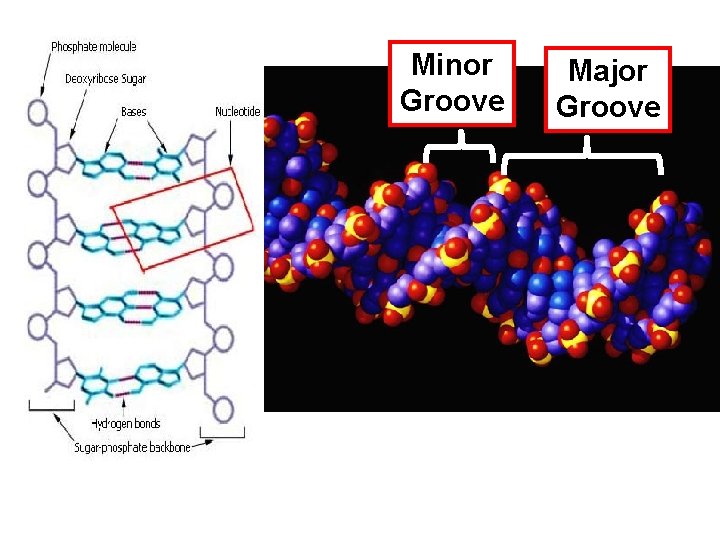
Minor Groove Major Groove

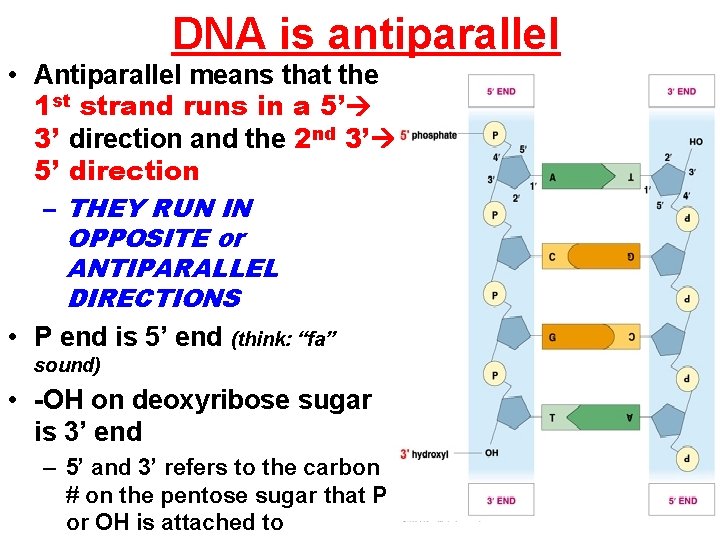
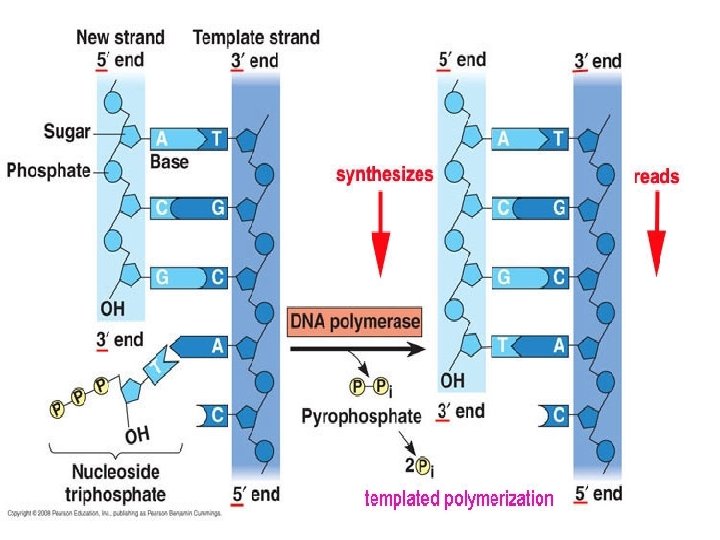
DNA is antiparallel • Antiparallel means that the 1 st strand runs in a 5’ 3’ direction and the 2 nd 3’ 5’ direction – THEY RUN IN OPPOSITE or ANTIPARALLEL DIRECTIONS • P end is 5’ end (think: “fa” sound) • -OH on deoxyribose sugar is 3’ end – 5’ and 3’ refers to the carbon # on the pentose sugar that P or OH is attached to

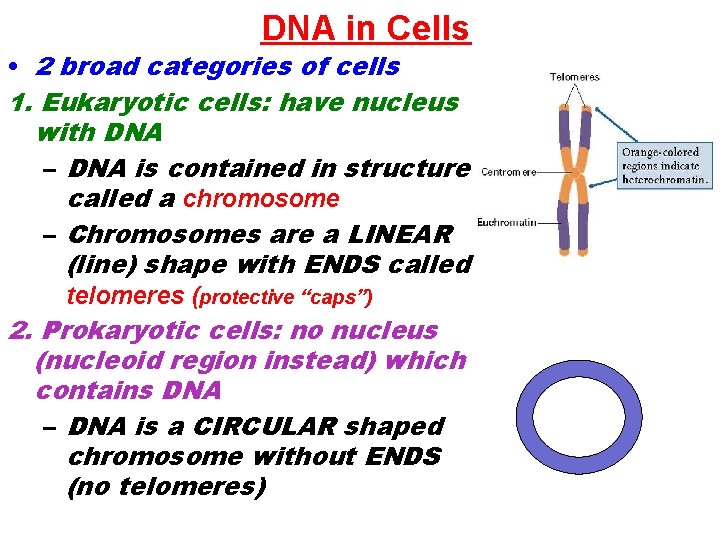
DNA in Cells • 2 broad categories of cells 1. Eukaryotic cells: have nucleus with DNA – DNA is contained in structure called a chromosome – Chromosomes are a LINEAR (line) shape with ENDS called telomeres (protective “caps”) 2. Prokaryotic cells: no nucleus (nucleoid region instead) which contains DNA – DNA is a CIRCULAR shaped chromosome without ENDS (no telomeres)

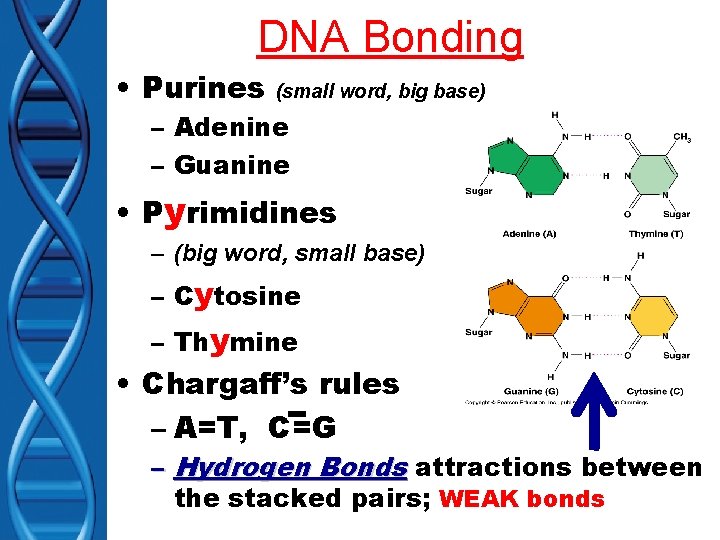
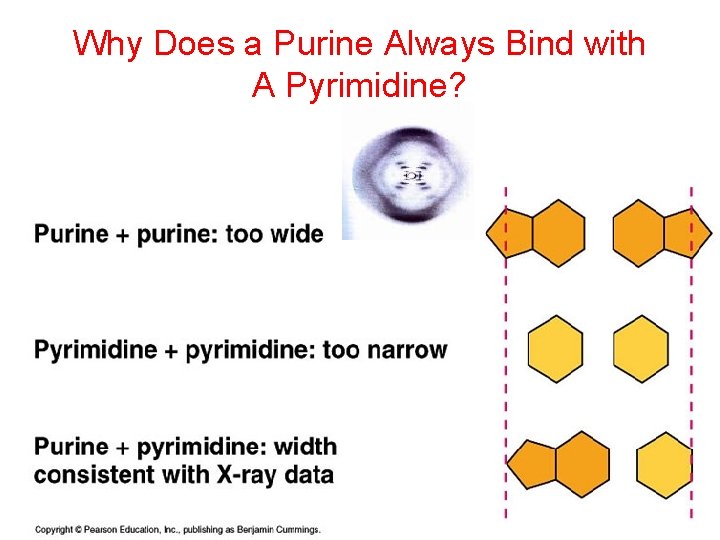
DNA Bonding • Purines (small word, big base) – Adenine – Guanine • Pyrimidines – (big word, small base) – Cytosine – Thymine • Chargaff’s rules – A=T, C=G – Hydrogen Bonds attractions between the stacked pairs; WEAK bonds

Why Does a Purine Always Bind with A Pyrimidine?

DNA Double Helix • http: //www. sumanasinc. com/webc ontent/animations/content/DNA_st ructure. html • Watson & Crick said that… – strands are complementary; nucleotides line up on template according to base pair rules (Chargaff’s rules) • A to T and C to G • LET’S PRACTICE… – Template: 5’ AATCGCTATAC 3’ – Complementary strand: 3’ TTAGCGATATG 5’

DNA Replication (Part 3 A-initiation)

DNA Replication • DNA Replication = DNA – Parent DNA makes 2 exact copies of DNA – Why? ? • Occurs in Cell Cycle before MITOSIS so each new cell can have its own FULL copy of DNA

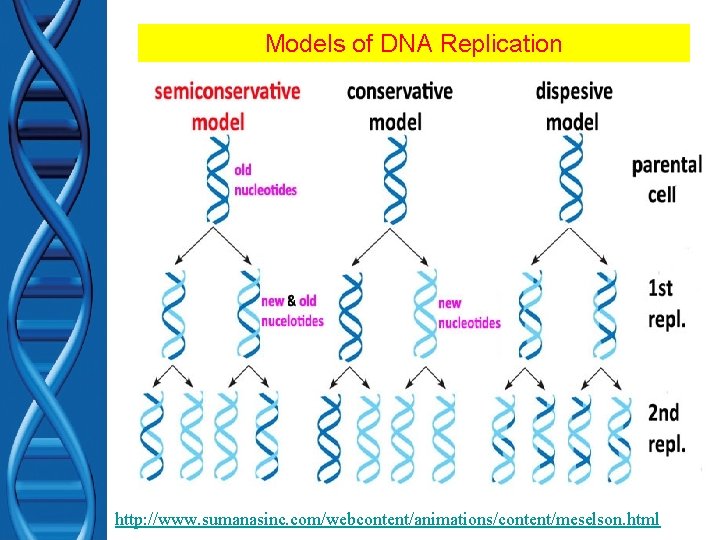
Models of DNA Replication http: //www. sumanasinc. com/webcontent/animations/content/meselson. html

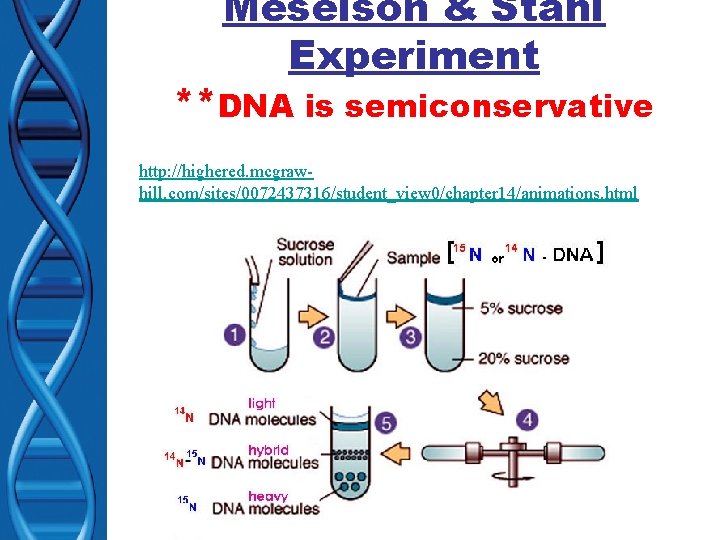
DNA Replication • How does it occur? • Matthew Meselson & Frank Stahl – Discovered replication is semiconservative – PROCEDURE varying densities of radioactive nitrogen (Nitrogen is in DNA)

Meselson & Stahl Experiment **DNA is semiconservative http: //highered. mcgrawhill. com/sites/0072437316/student_view 0/chapter 14/animations. html

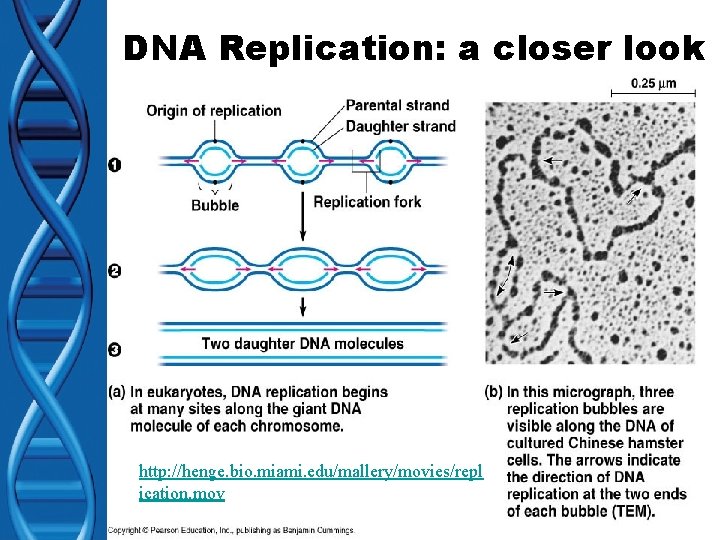
DNA Replication: a closer look http: //henge. bio. miami. edu/mallery/movies/repl ication. mov

DNA Replication Steps: • Initiation – involves assembly of replication fork (bubble) at origin of replication • sequence of DNA found at a specific site • Elongation – Parental strands unwind and daughter strands are synthesized. – the addition of bases by proteins • Termination: – the duplicated chromosomes separate from each other. Now, there are 2 IDENTICAL copies of DNA.

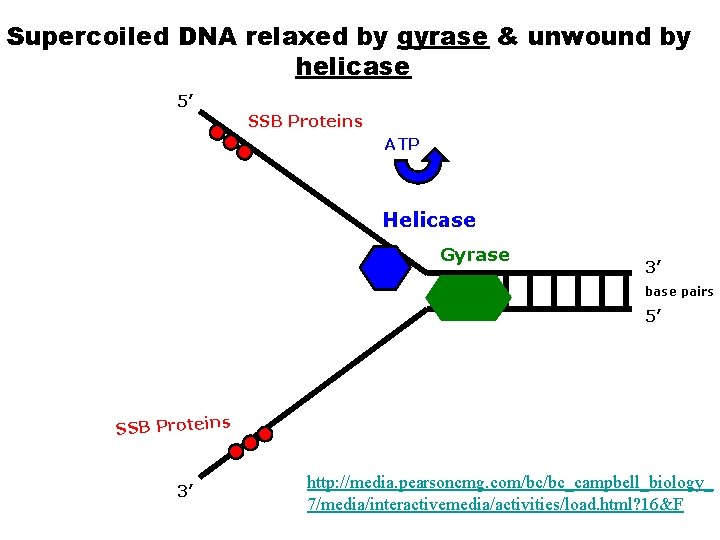
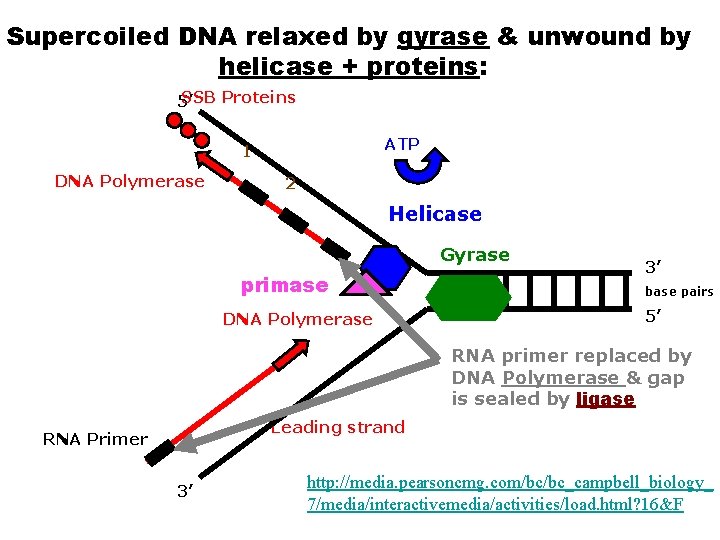
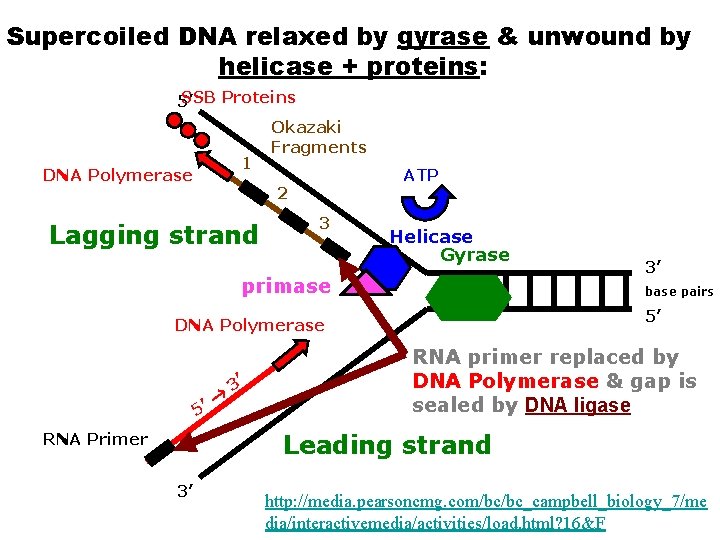
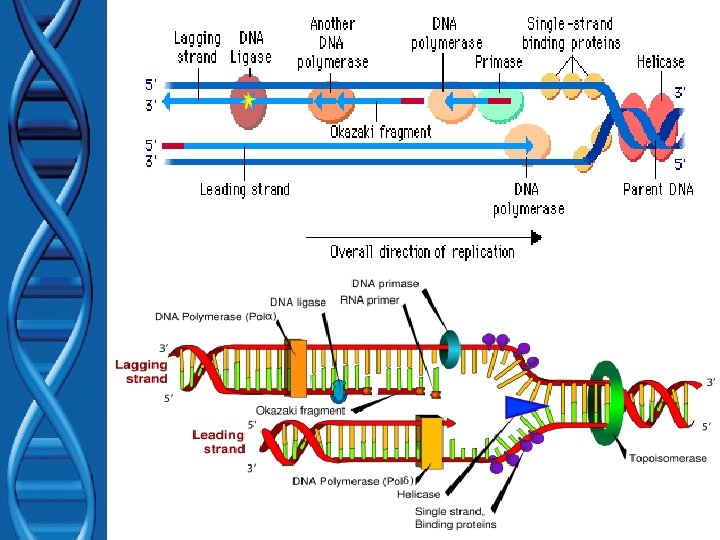
Segments of single-stranded DNA are called template strands. Copied strand is called the complement strand (think “c” for copy) BEGINNING OF DNA REPLICATION (INITIATION) • Gyrase (type of topoisomerase) – relaxes the supercoiled DNA. • DNA helicase (think “helix”) – binds to the DNA at the replication fork – untwist (“unzips”) DNA using energy from ATP – Breaks hydrogen bonds between base pairs • Single-stranded DNA-binding proteins (SSBP) – stabilize the single-stranded template DNA during the process so they don’t bond back together.

Supercoiled DNA relaxed by gyrase & unwound by helicase 5’ SSB Proteins ATP Helicase Gyrase 3’ base pairs 5’ SSB Proteins 3’ http: //media. pearsoncmg. com/bc/bc_campbell_biology_ 7/media/interactivemedia/activities/load. html? 16&F

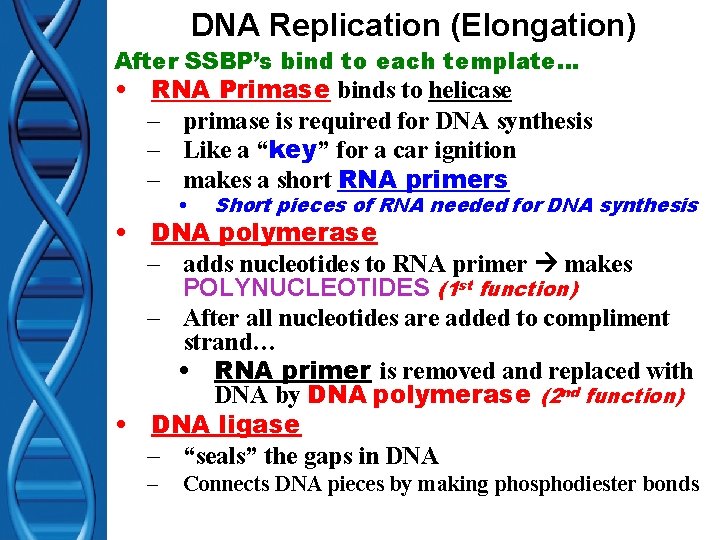
DNA Replication (Elongation) After SSBP’s bind to each template… • RNA Primase binds to helicase – primase is required for DNA synthesis – Like a “key” for a car ignition – makes a short RNA primers • Short pieces of RNA needed for DNA synthesis • DNA polymerase – adds nucleotides to RNA primer makes POLYNUCLEOTIDES (1 st function) – After all nucleotides are added to compliment strand… • RNA primer is removed and replaced with DNA by DNA polymerase (2 nd function) • DNA ligase – “seals” the gaps in DNA – Connects DNA pieces by making phosphodiester bonds

Supercoiled DNA relaxed by gyrase & unwound by helicase + proteins: SSB Proteins 5’ ATP 1 DNA Polymerase 2 Helicase Gyrase primase DNA Polymerase 3’ base pairs 5’ RNA primer replaced by DNA Polymerase & gap is sealed by ligase Leading strand RNA Primer 3’ http: //media. pearsoncmg. com/bc/bc_campbell_biology_ 7/media/interactivemedia/activities/load. html? 16&F

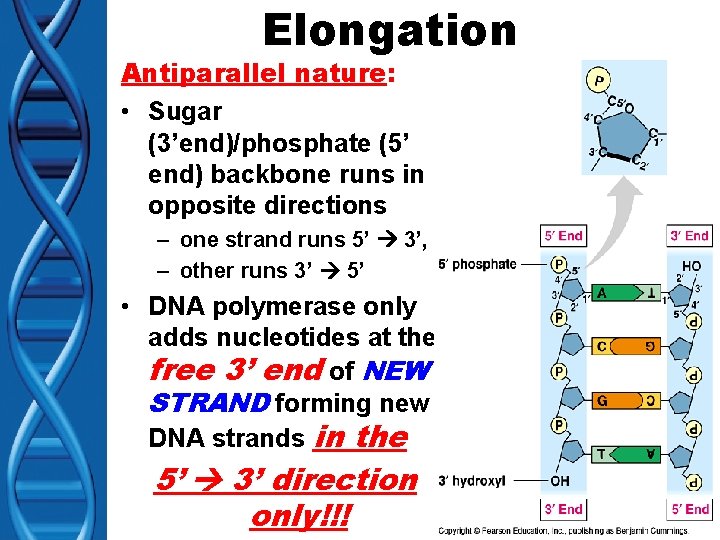
Elongation Antiparallel nature: • Sugar (3’end)/phosphate (5’ end) backbone runs in opposite directions – one strand runs 5’ 3’, – other runs 3’ 5’ • DNA polymerase only adds nucleotides at the free 3’ end of NEW STRAND forming new DNA strands in the 5’ 3’ direction only!!!


Elongation (con’t) • Leading (daughter) strand – NEW strand made toward the replication fork (only in 5’ 3’ direction from the 3’ 5’ master strand – Needs ONE (1) RNA primer made by Primase – This new leading strand is made CONTINOUSLY

Elongation (con’t) Lagging (daughter) strand • NEW strand synthesis away from replication fork • Replicate DISCONTINUOUSLY – Creates Okazaki fragments • Short pieces of DNA – Okazaki fragments joined by DNA ligase • “Stitches” fragments together – Needs MANY RNA primer made by Primase •

Supercoiled DNA relaxed by gyrase & unwound by helicase + proteins: SSB Proteins 5’ 1 DNA Polymerase Okazaki Fragments ATP 2 Lagging strand 3 Helicase Gyrase primase base pairs 5’ DNA Polymerase 5’ 3’ 3’ RNA primer replaced by DNA Polymerase & gap is sealed by DNA ligase Leading strand RNA Primer 3’ http: //media. pearsoncmg. com/bc/bc_campbell_biology_7/me dia/interactivemedia/activities/load. html? 16&F


DNA Replication: Elongation http: //highered. mcgrawhill. com/sites/0072437316/student_view 0/chapter 14/animations. html

DNA Replication (Part 3 C-termination)

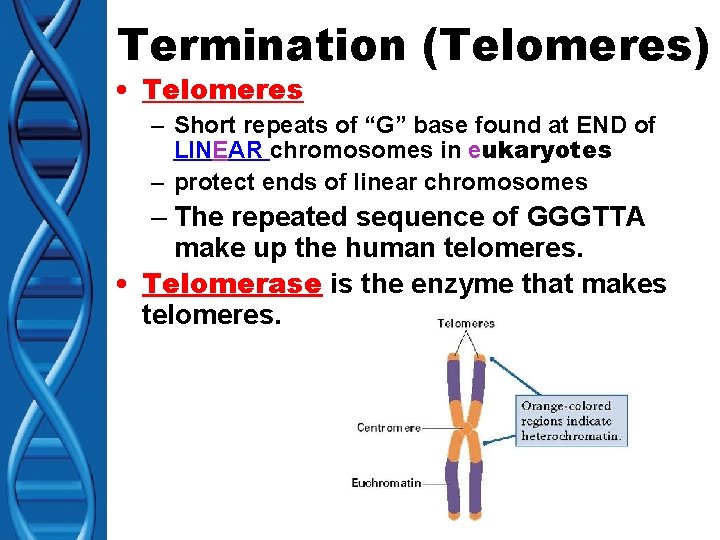
Termination (Telomeres) • Telomeres – Short repeats of “G” base found at END of LINEAR chromosomes in eukaryotes – protect ends of linear chromosomes – The repeated sequence of GGGTTA make up the human telomeres. • Telomerase is the enzyme that makes telomeres.

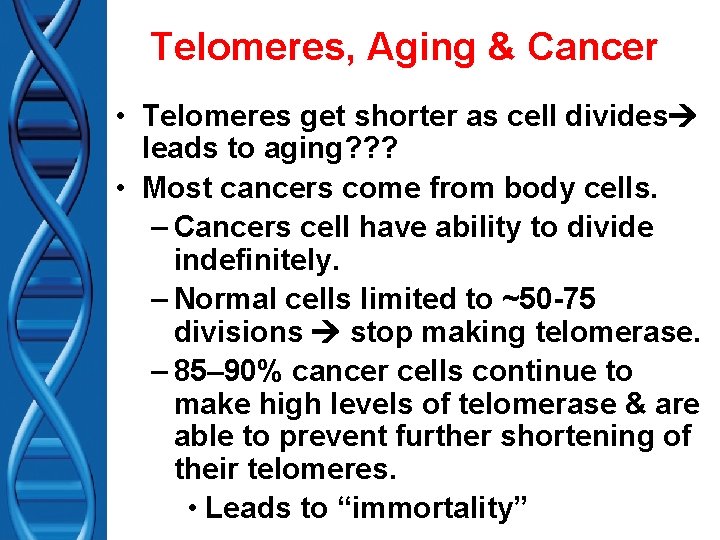
Telomeres, Aging & Cancer • Telomeres get shorter as cell divides leads to aging? ? ? • Most cancers come from body cells. – Cancers cell have ability to divide indefinitely. – Normal cells limited to ~50 -75 divisions stop making telomerase. – 85– 90% cancer cells continue to make high levels of telomerase & are able to prevent further shortening of their telomeres. • Leads to “immortality”

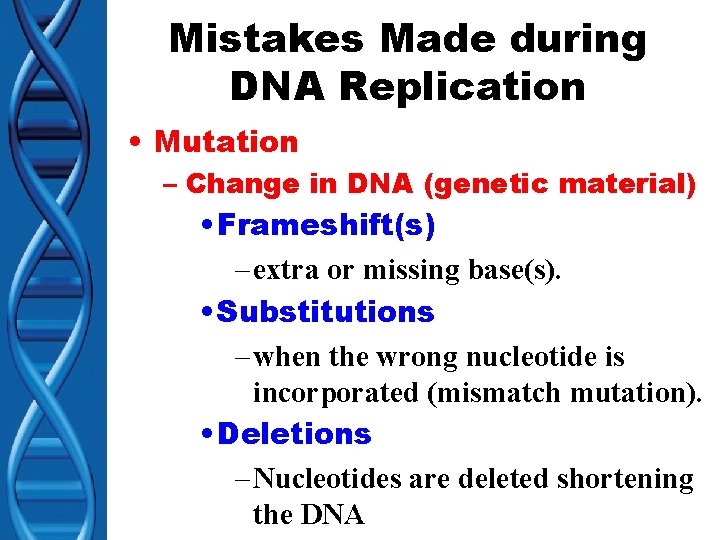
Mistakes Made during DNA Replication • Mutation – Change in DNA (genetic material) • Frameshift(s) – extra or missing base(s). • Substitutions – when the wrong nucleotide is incorporated (mismatch mutation). • Deletions – Nucleotides are deleted shortening the DNA

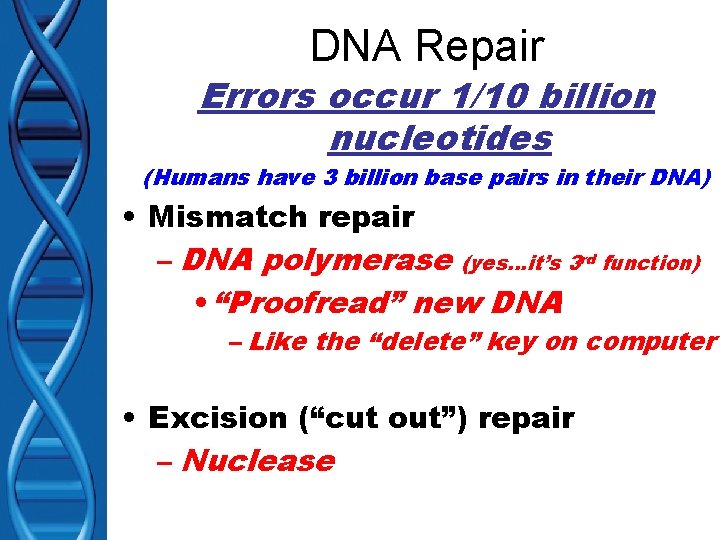
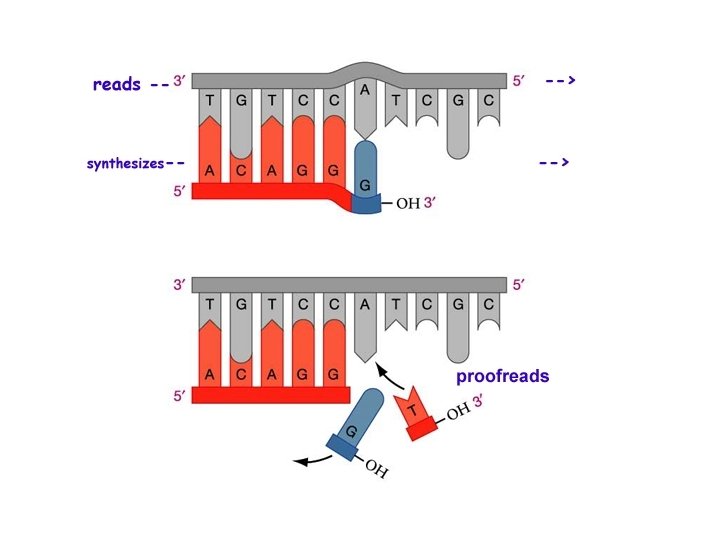
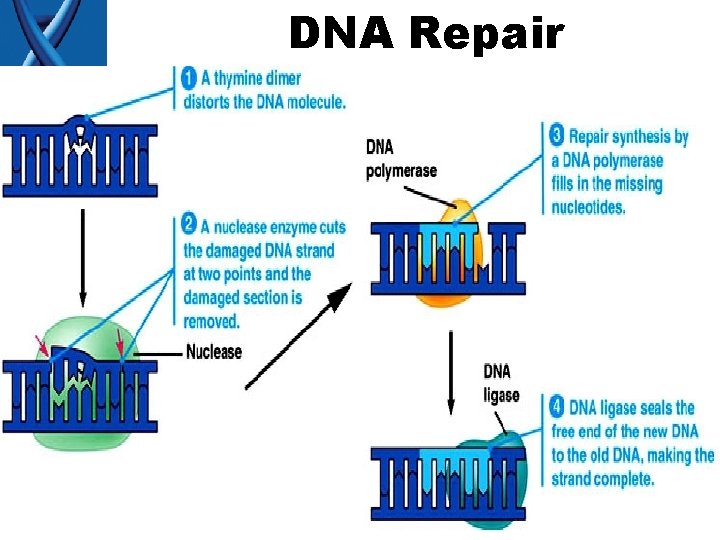
DNA Repair Errors occur 1/10 billion nucleotides (Humans have 3 billion base pairs in their DNA) • Mismatch repair – DNA polymerase (yes…it’s 3 rd function) • “Proofread” new DNA – Like the “delete” key on computer • Excision (“cut out”) repair – Nuclease


DNA Repair