Chapter 11 Statistical Interpretation Fundamentals of Forensic DNA



- Slides: 14

Chapter 11 Statistical Interpretation Fundamentals of Forensic DNA Typing Slides prepared by John M. Butler June 2009

Chapter 11 – Statistical Data Interpretation Chapter Summary Matching DNA results must be provided with statistical interpretation to help determine their relevance. The frequency of alleles and genotypes are assessed by gathering a sampling of a particular population. Provided that the alleles and their loci are independent from one another, results can be combined using what is commonly referred to as “the product rule. ” The random match probability for a particular DNA profile represents the chance of drawing this combination of alleles at random from a population of unrelated individuals and is not the probability of guilt—a philosophical mistake known as the “prosecutor’s fallacy. ” Corrections for subpopulation structure and possible involvement of relatives reduce the match probability and typically provide a more conservative estimate for the defendant. Unresolved mixtures and partial profiles, which are forensic realities, reduce the match probability for a particular sample.

DNA Testing Requires a Reference Sample A DNA profile by itself is fairly useless because it has no context… DNA analysis for identity only works by comparison – you need a reference sample Crime Scene Evidence compared to Suspect(s) (Forensic Case) Child compared to Alleged Father (Paternity Case) Victim’s Remains compared to Biological Relative (Mass Disaster ID) Soldier’s Remains compared to Direct Reference Sample (Armed Forces ID)

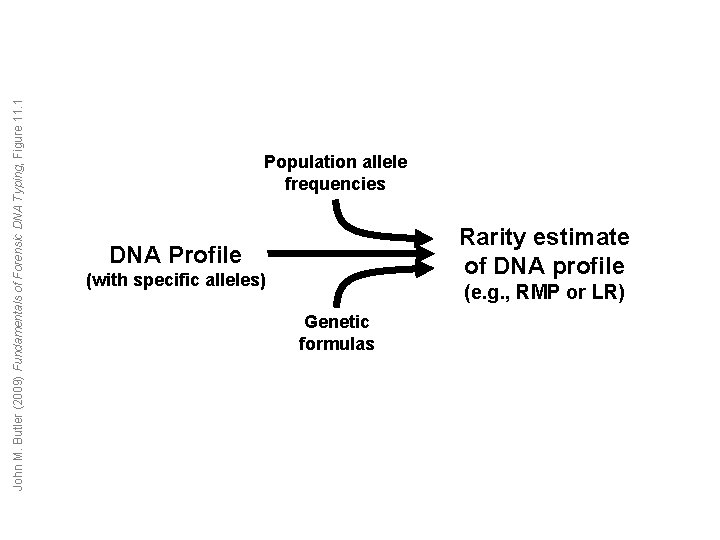
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 11. 1 Population allele frequencies Rarity estimate of DNA profile DNA Profile (with specific alleles) (e. g. , RMP or LR) Genetic formulas

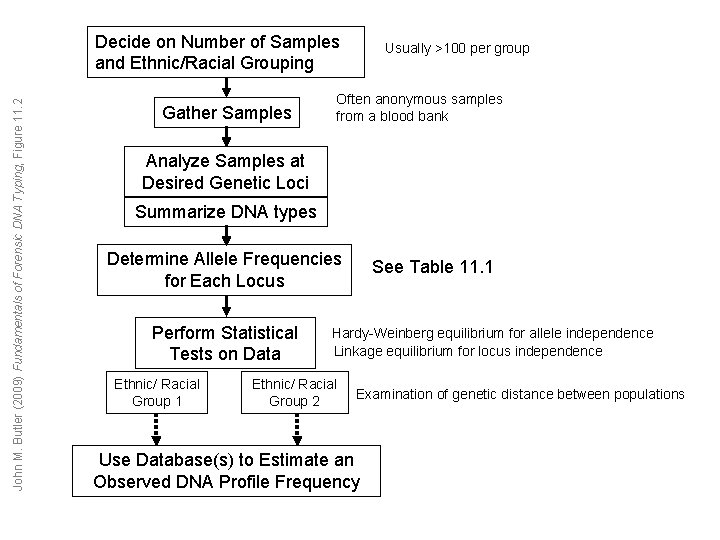
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 11. 2 Decide on Number of Samples and Ethnic/Racial Grouping Gather Samples Usually >100 per group Often anonymous samples from a blood bank Analyze Samples at Desired Genetic Loci Summarize DNA types Determine Allele Frequencies for Each Locus Perform Statistical Tests on Data Ethnic/ Racial Group 1 See Table 11. 1 Hardy-Weinberg equilibrium for allele independence Linkage equilibrium for locus independence Ethnic/ Racial Group 2 Examination of genetic distance between populations Use Database(s) to Estimate an Observed DNA Profile Frequency

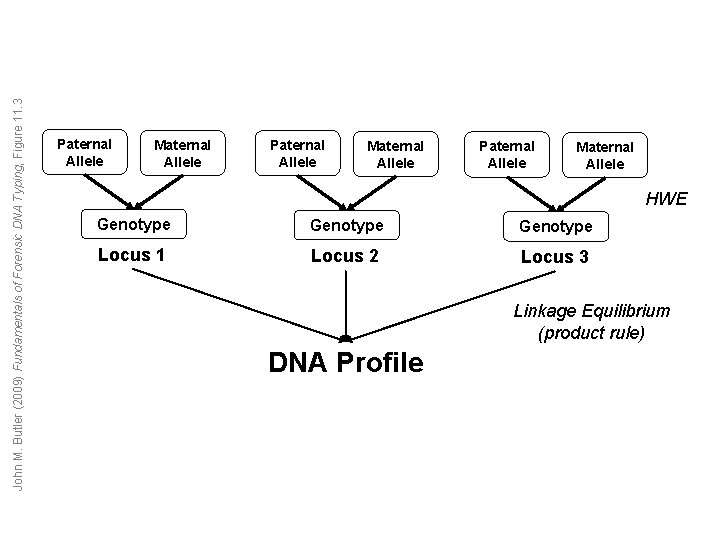
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 11. 3 Paternal Allele Maternal Allele HWE Genotype Locus 1 Locus 2 Locus 3 Linkage Equilibrium (product rule) DNA Profile

How Statistical Calculations are Made • Generate data with set(s) of samples from desired population group(s) – Generally only 100 -150 samples are needed to obtain reliable allele frequency estimates • Determine allele frequencies at each locus – Count number of each allele seen • Allele frequency information is used to estimate the rarity of a particular DNA profile – Homozygotes (p 2), Heterozygotes (2 pq) – Product rule used (multiply locus frequency estimates)

How Are Such Large Numbers Generated with Random Match Probabilities? • Each allele is sampled multiple times to produce a statistically stable allele frequency • Using theoretical model from genetics, multiple loci are multiplied together to produce an estimate of the rarity of a particular DNA profile (combination of STR alleles based on individual allele frequencies) • Remember that relatives will share genetic characteristics and thus have STR profiles that are more similar to one another than unrelated individuals • We are not looking at every person on the planet nor are we looking at every nucleotide in the suspect’s genome

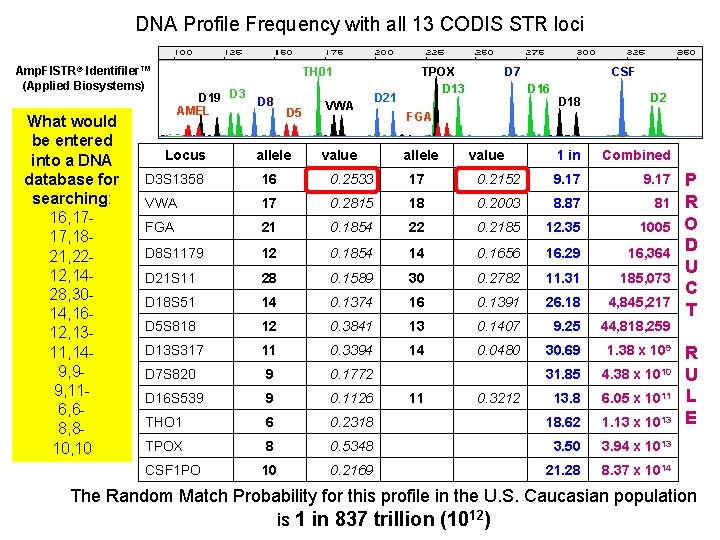
DNA Profile Frequency with all 13 CODIS STR loci Amp. Fl. STR® Identifiler™ (Applied Biosystems) What would be entered into a DNA database for searching: 16, 1717, 1821, 2212, 1428, 3014, 1612, 1311, 149, 99, 116, 68, 810, 10 TH 01 D 19 D 3 AMEL Locus D 8 D 5 allele VWA value D 21 TPOX D 13 D 7 CSF D 16 D 18 D 2 FGA allele value 1 in Combined D 3 S 1358 16 0. 2533 17 0. 2152 9. 17 VWA 17 0. 2815 18 0. 2003 8. 87 81 FGA 21 0. 1854 22 0. 2185 12. 35 1005 D 8 S 1179 12 0. 1854 14 0. 1656 16. 29 16, 364 D 21 S 11 28 0. 1589 30 0. 2782 11. 31 185, 073 D 18 S 51 14 0. 1374 16 0. 1391 26. 18 4, 845, 217 D 5 S 818 12 0. 3841 13 0. 1407 9. 25 44, 818, 259 D 13 S 317 11 0. 3394 14 0. 0480 30. 69 1. 38 x 109 D 7 S 820 9 0. 1772 31. 85 4. 38 x 1010 D 16 S 539 9 0. 1126 13. 8 6. 05 x 1011 THO 1 6 0. 2318 18. 62 1. 13 x 1013 TPOX 8 0. 5348 3. 50 3. 94 x 1013 CSF 1 PO 10 0. 2169 21. 28 8. 37 x 1014 11 0. 3212 P R O D U C T R U L E The Random Match Probability for this profile in the U. S. Caucasian population is 1 in 837 trillion (1012)

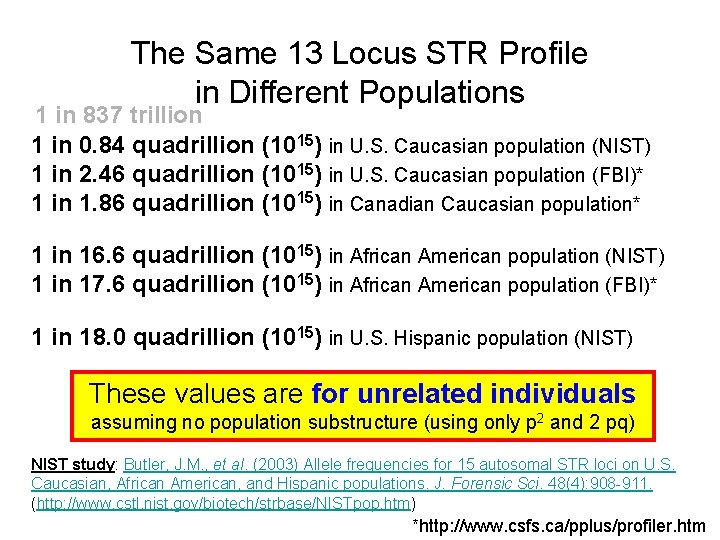
The Same 13 Locus STR Profile in Different Populations 1 in 837 trillion 1 in 0. 84 quadrillion (1015) in U. S. Caucasian population (NIST) 1 in 2. 46 quadrillion (1015) in U. S. Caucasian population (FBI)* 1 in 1. 86 quadrillion (1015) in Canadian Caucasian population* 1 in 16. 6 quadrillion (1015) in African American population (NIST) 1 in 17. 6 quadrillion (1015) in African American population (FBI)* 1 in 18. 0 quadrillion (1015) in U. S. Hispanic population (NIST) These values are for unrelated individuals assuming no population substructure (using only p 2 and 2 pq) NIST study: Butler, J. M. , et al. (2003) Allele frequencies for 15 autosomal STR loci on U. S. Caucasian, African American, and Hispanic populations. J. Forensic Sci. 48(4): 908 -911. (http: //www. cstl. nist. gov/biotech/strbase/NISTpop. htm) *http: //www. csfs. ca/pplus/profiler. htm

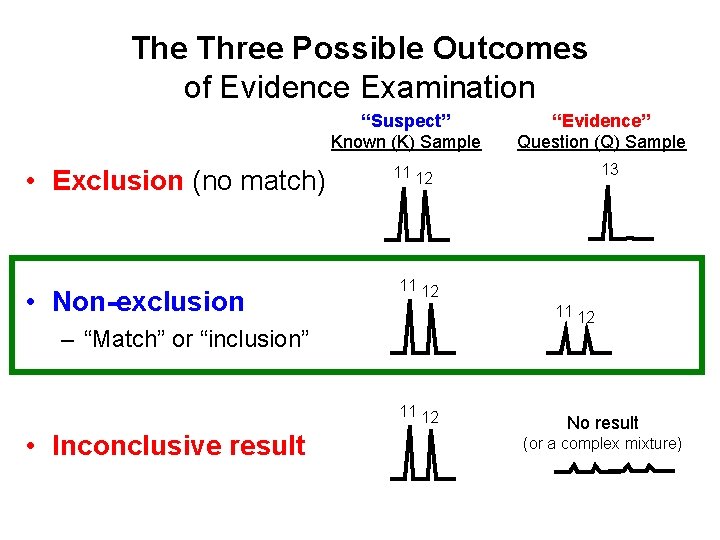
The Three Possible Outcomes of Evidence Examination “Suspect” Known (K) Sample • Exclusion (no match) • Non-exclusion 13 11 12 – “Match” or “inclusion” 11 12 • Inconclusive result “Evidence” Question (Q) Sample No result (or a complex mixture)

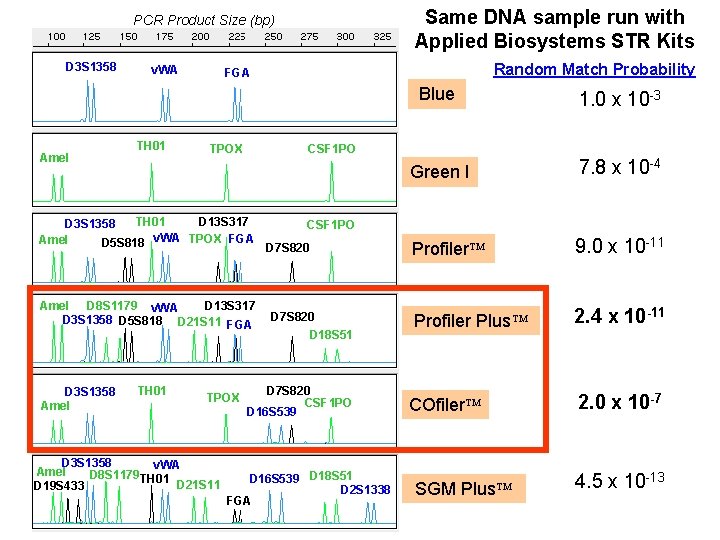
Same DNA sample run with Applied Biosystems STR Kits PCR Product Size (bp) D 3 S 1358 Amel v. WA TH 01 D 13 S 317 Amel D 8 S 1179 v. WA D 3 S 1358 D 5 S 818 D 21 S 11 FGA TH 01 TPOX D 3 S 1358 v. WA Amel D 8 S 1179 TH 01 D 21 S 11 D 19 S 433 Blue 1. 0 x 10 -3 Green I 7. 8 x 10 -4 Profiler 9. 0 x 10 -11 Profiler Plus 2. 4 x 10 -11 COfiler 2. 0 x 10 -7 CSF 1 PO TPOX TH 01 D 13 S 317 D 3 S 1358 Amel D 5 S 818 v. WA TPOX FGA D 3 S 1358 Amel Random Match Probability FGA CSF 1 PO D 7 S 820 D 18 S 51 D 7 S 820 CSF 1 PO D 16 S 539 D 18 S 51 D 2 S 1338 FGA SGM Plus 4. 5 x 10 -13

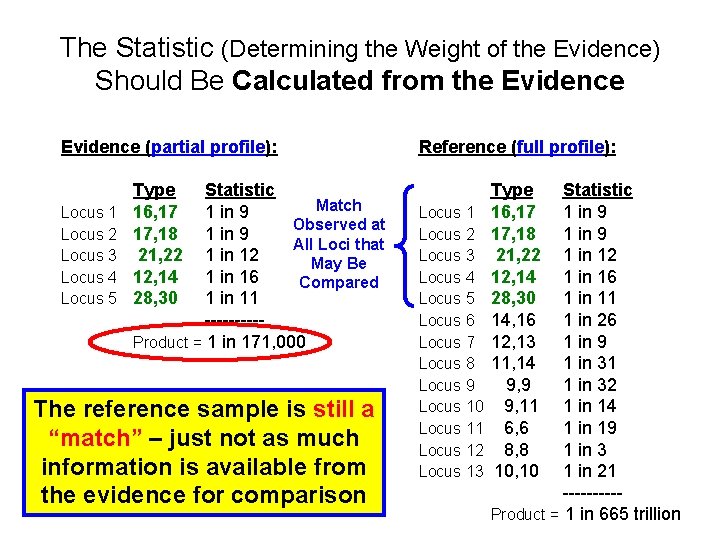
The Statistic (Determining the Weight of the Evidence) Should Be Calculated from the Evidence (partial profile): Locus 1 Locus 2 Locus 3 Locus 4 Locus 5 Type 16, 17 17, 18 21, 22 12, 14 28, 30 Statistic Match 1 in 9 Observed at 1 in 9 All Loci that 1 in 12 May Be 1 in 16 Compared 1 in 11 -----Product = 1 in 171, 000 The reference sample is still a “match” – just not as much information is available from the evidence for comparison Reference (full profile): Locus 1 Locus 2 Locus 3 Locus 4 Locus 5 Locus 6 Locus 7 Locus 8 Locus 9 Locus 10 Locus 11 Locus 12 Locus 13 Type 16, 17 17, 18 21, 22 12, 14 28, 30 14, 16 12, 13 11, 14 9, 9 9, 11 6, 6 8, 8 10, 10 Statistic 1 in 9 1 in 12 1 in 16 1 in 11 1 in 26 1 in 9 1 in 31 1 in 32 1 in 14 1 in 19 1 in 3 1 in 21 -----Product = 1 in 665 trillion

Chapter 11 – Points for Discussion • What is the purpose of providing a random match probability statistic when two DNA profiles match? • What is the purpose of generating a population database? • For a locus with n possible alleles, how many total genotypes are theoretically possible? • Why utilize a minimum allele frequency? • Why is it important to establish independence between alleles and between loci? • What is wrong with simply saying that a suspect is included in a mixture without providing any statistics?