Chapter 11 Membrane Transport of Small Molecules and
Chapter 11 • Membrane Transport of Small Molecules and the Electrical Properties of Membranes

I. Principles of membrane transport - protein-free lipid bilayers are highly impermeable to ions - two main classes of membrane transport proteins - transporters ( also called carriers or permeases) and channels - active transport – mediated by transporters coupled to an energy source - passive transport – mediated by all channels and many transporters II. Transporters and passive transport - glucose transporters III. Transporters and active transport - coupled transporters; ATP-driven pumps; light-driven pumps - uniporters; symporters; antiporters - Na+-glucose cotransporter – coupled transport; symporter - lactose permease - coupled transport; symporter - three classes of ATP-driven pumps - Ca 2+ pump - Na+-K+ pump - ABC transporters IV. Ion channels - ion channels are selective and gated - membrane potential - K+- channel - aquaporins - neuron - voltage-gated Na+-channels - transmitter-gated ion channels - neuromuscular transmission

Principles of membrane transport
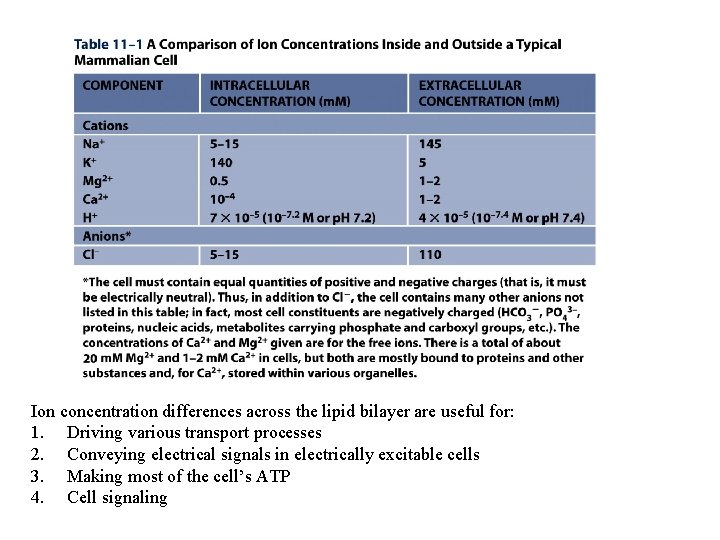
Ion concentration differences across the lipid bilayer are useful for: 1. Driving various transport processes 2. Conveying electrical signals in electrically excitable cells 3. Making most of the cell’s ATP 4. Cell signaling
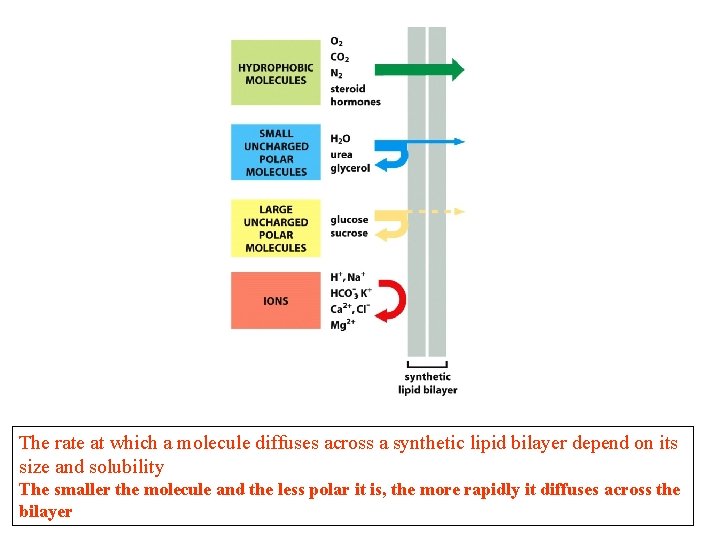
The rate at which a molecule diffuses across a synthetic lipid bilayer depend on its size and solubility The smaller the molecule and the less polar it is, the more rapidly it diffuses across the bilayer
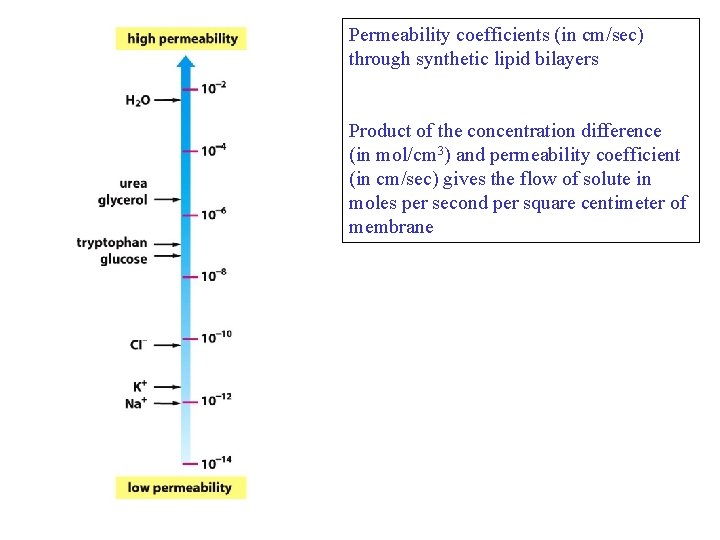
Permeability coefficients (in cm/sec) through synthetic lipid bilayers Product of the concentration difference (in mol/cm 3) and permeability coefficient (in cm/sec) gives the flow of solute in moles per second per square centimeter of membrane
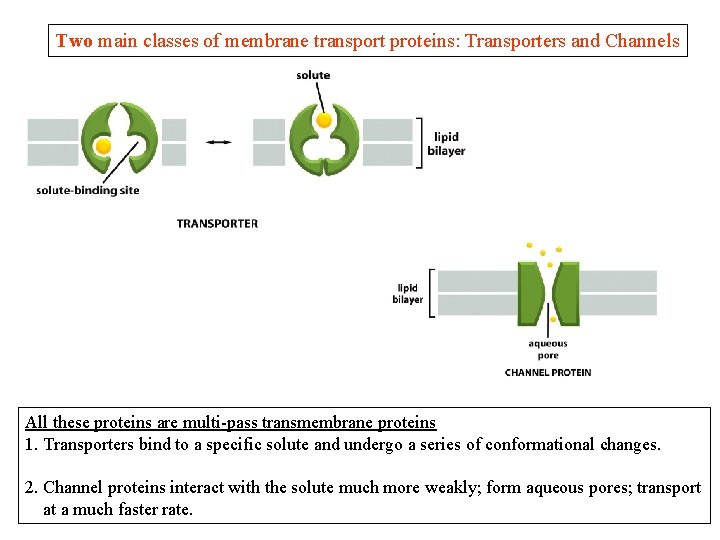
Two main classes of membrane transport proteins: Transporters and Channels All these proteins are multi-pass transmembrane proteins 1. Transporters bind to a specific solute and undergo a series of conformational changes. 2. Channel proteins interact with the solute much more weakly; form aqueous pores; transport at a much faster rate.
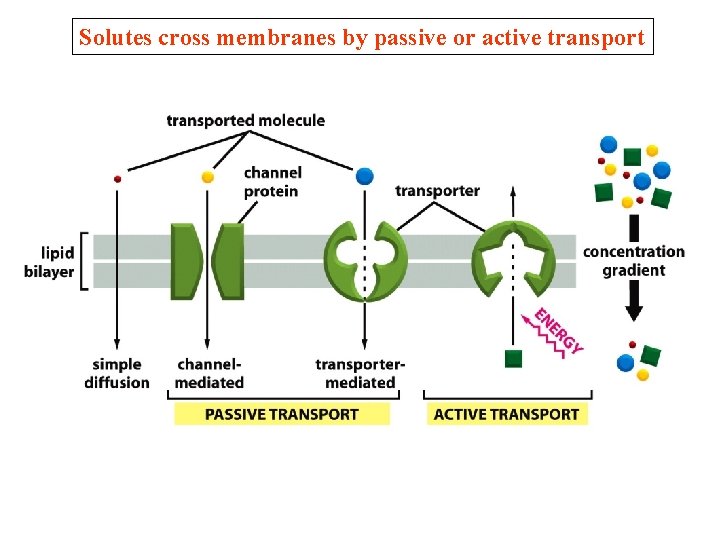
Solutes cross membranes by passive or active transport
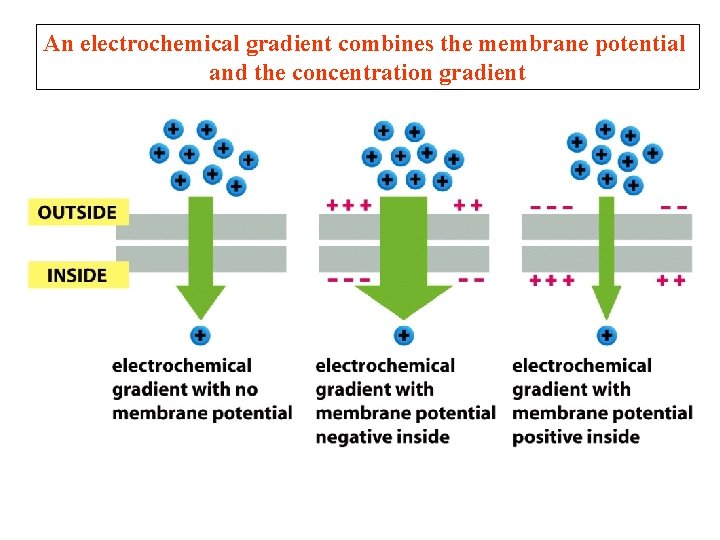
An electrochemical gradient combines the membrane potential and the concentration gradient
Transporters and passive transport
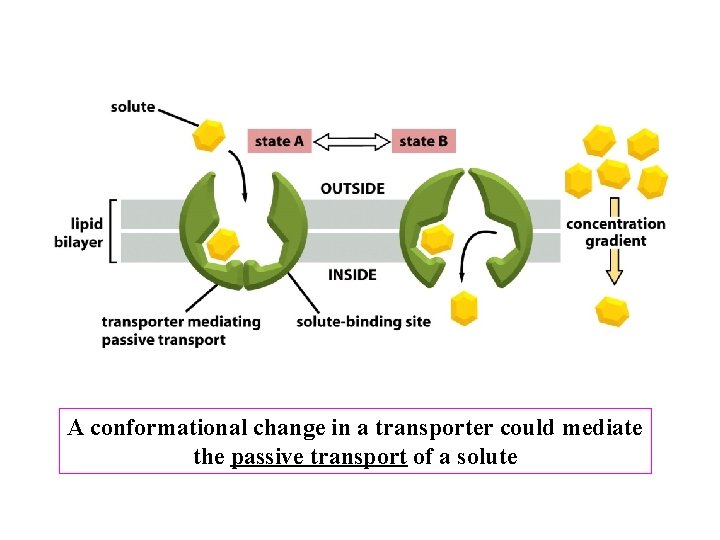
A conformational change in a transporter could mediate the passive transport of a solute
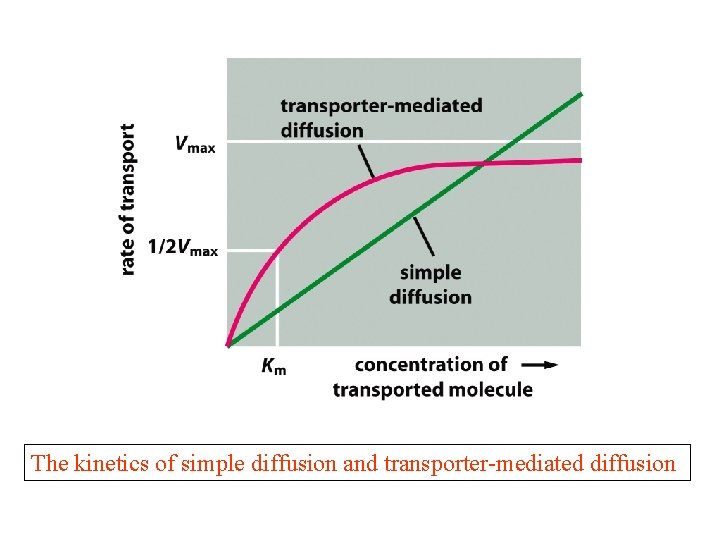
The kinetics of simple diffusion and transporter-mediated diffusion
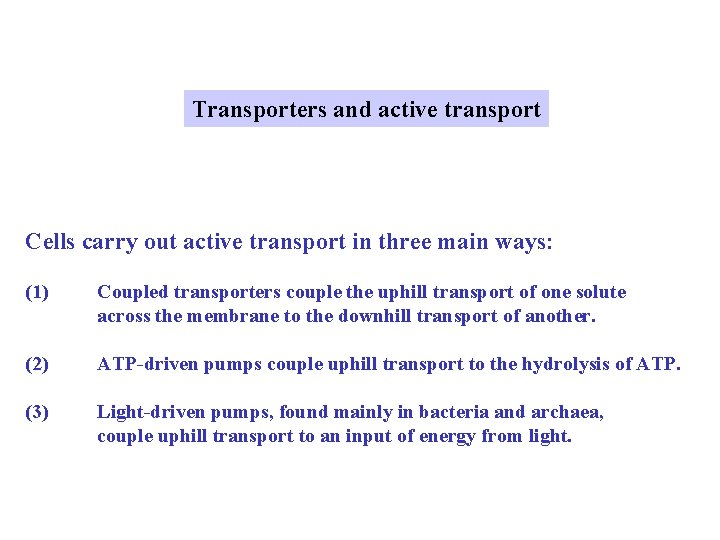
Transporters and active transport Cells carry out active transport in three main ways: (1) Coupled transporters couple the uphill transport of one solute across the membrane to the downhill transport of another. (2) ATP-driven pumps couple uphill transport to the hydrolysis of ATP. (3) Light-driven pumps, found mainly in bacteria and archaea, couple uphill transport to an input of energy from light.
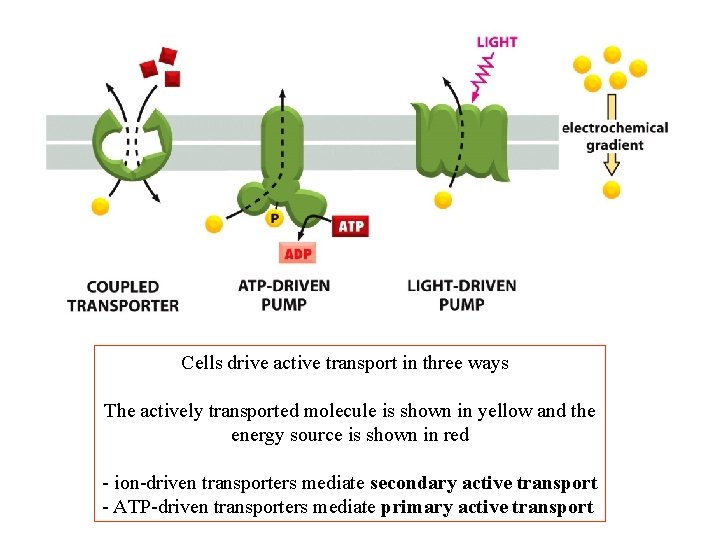
Cells drive active transport in three ways The actively transported molecule is shown in yellow and the energy source is shown in red - ion-driven transporters mediate secondary active transport - ATP-driven transporters mediate primary active transport
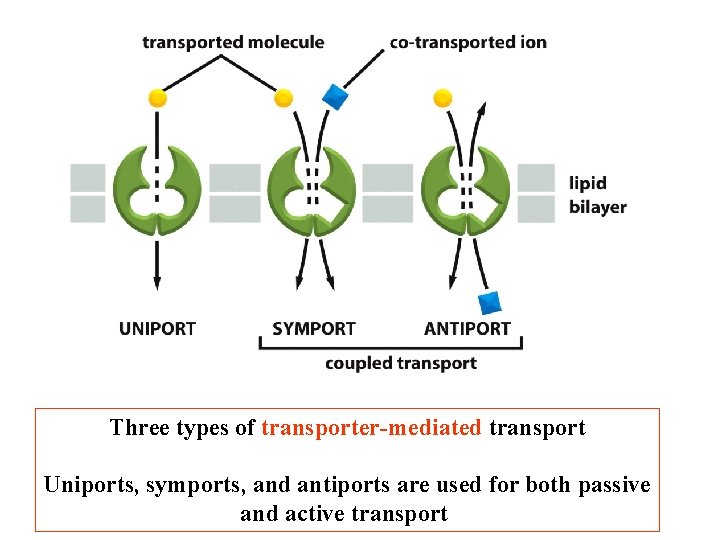
Three types of transporter-mediated transport Uniports, symports, and antiports are used for both passive and active transport
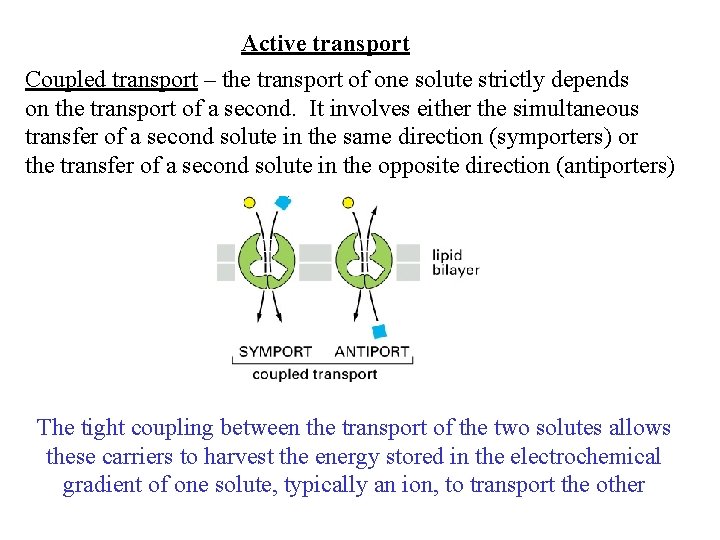
Active transport Coupled transport – the transport of one solute strictly depends on the transport of a second. It involves either the simultaneous transfer of a second solute in the same direction (symporters) or the transfer of a second solute in the opposite direction (antiporters) The tight coupling between the transport of the two solutes allows these carriers to harvest the energy stored in the electrochemical gradient of one solute, typically an ion, to transport the other

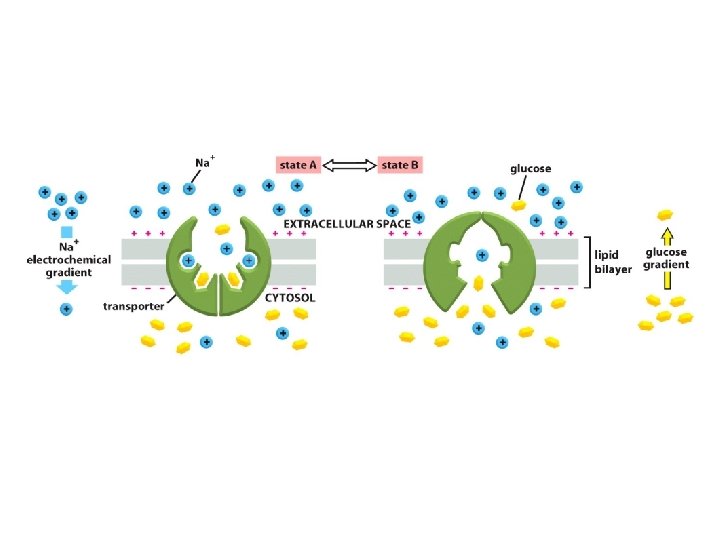
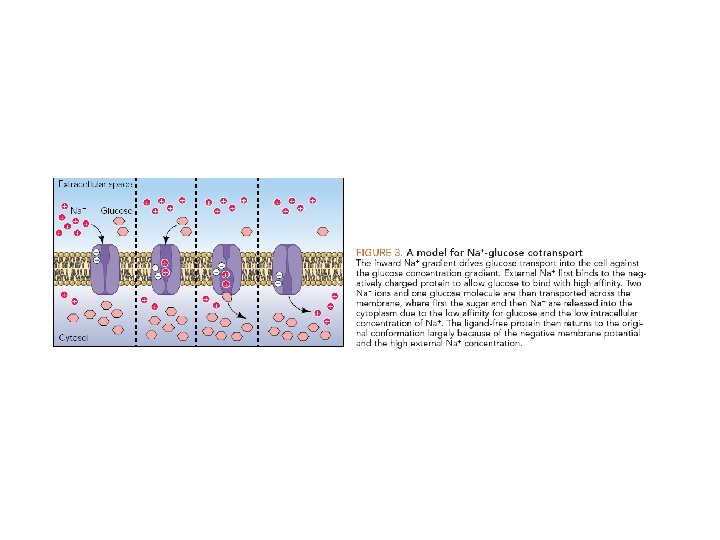
The glucose-Na+ symport protein uses the electrochemical Na+ gradient to drive the import of glucose
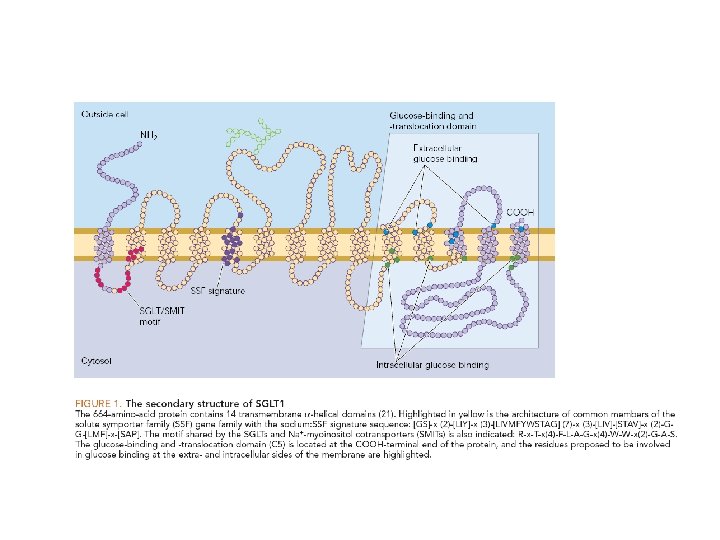
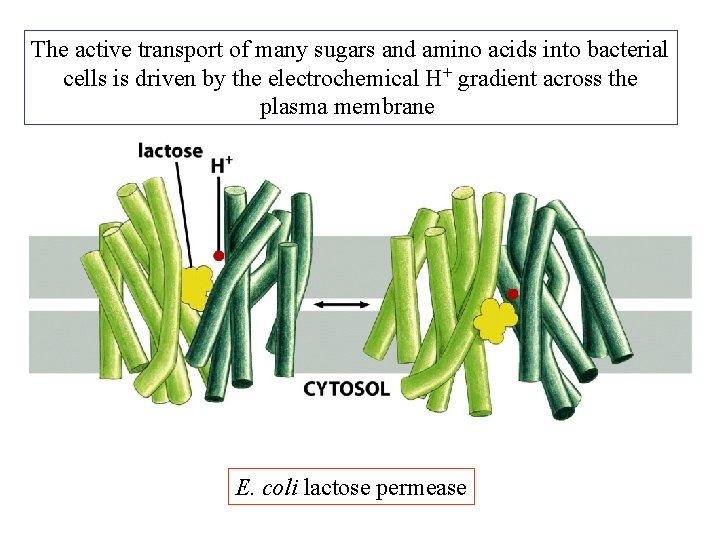
The active transport of many sugars and amino acids into bacterial cells is driven by the electrochemical H+ gradient across the plasma membrane E. coli lactose permease
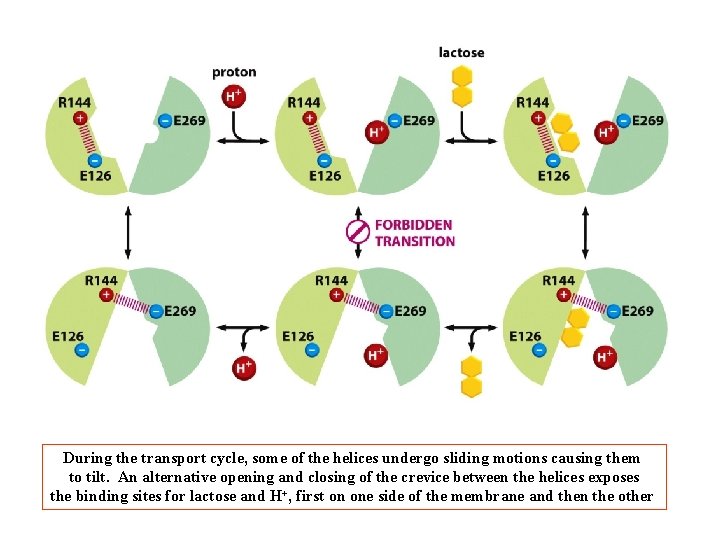
During the transport cycle, some of the helices undergo sliding motions causing them to tilt. An alternative opening and closing of the crevice between the helices exposes the binding sites for lactose and H+, first on one side of the membrane and then the other
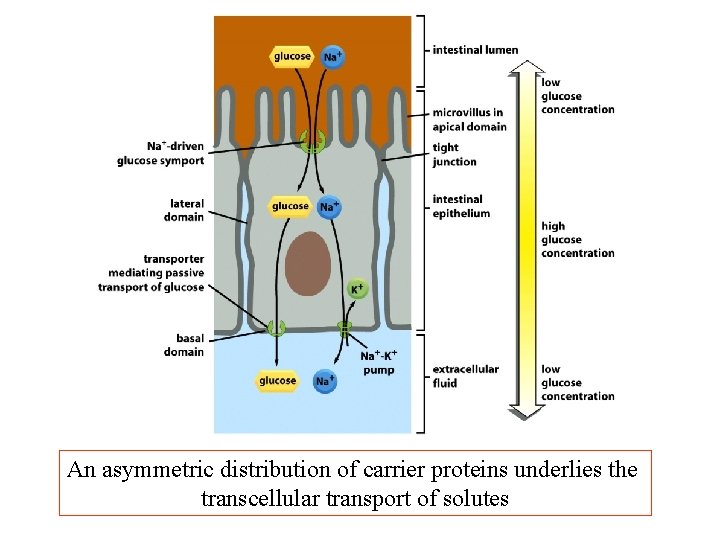
An asymmetric distribution of carrier proteins underlies the transcellular transport of solutes
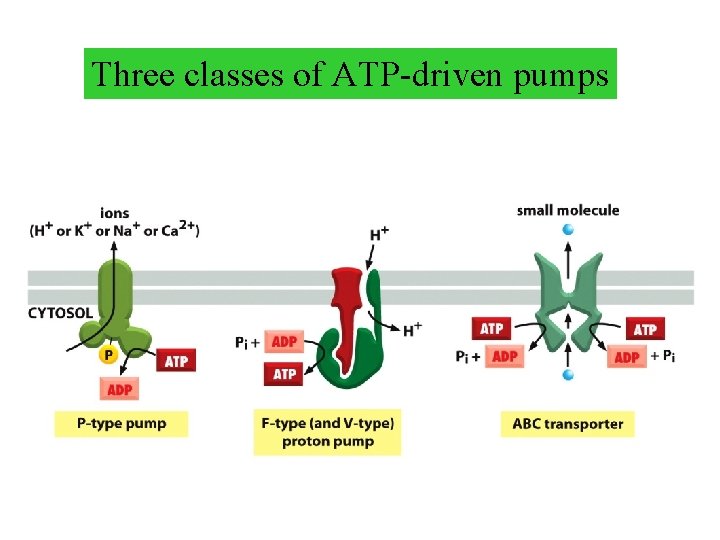
Three classes of ATP-driven pumps
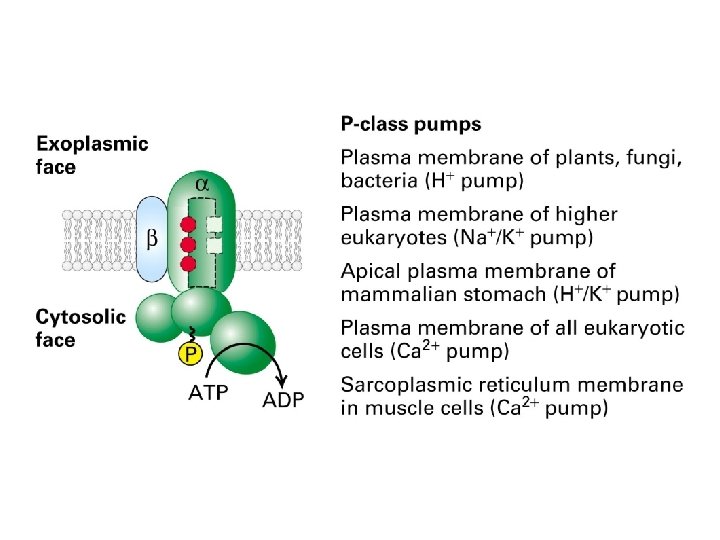
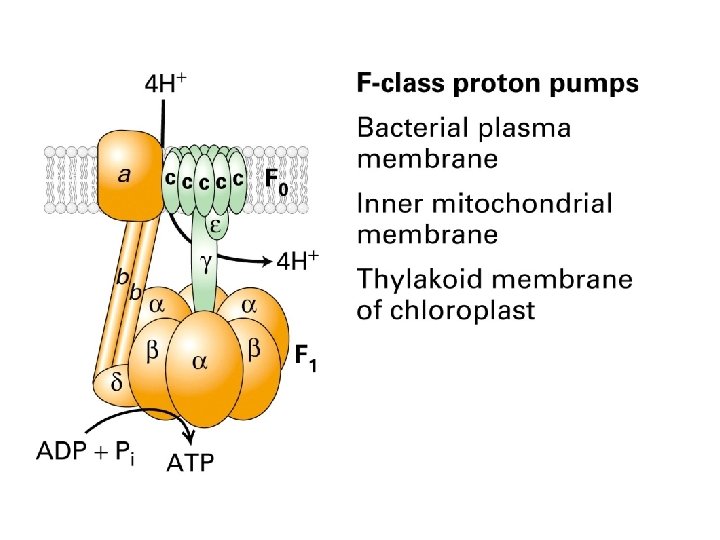
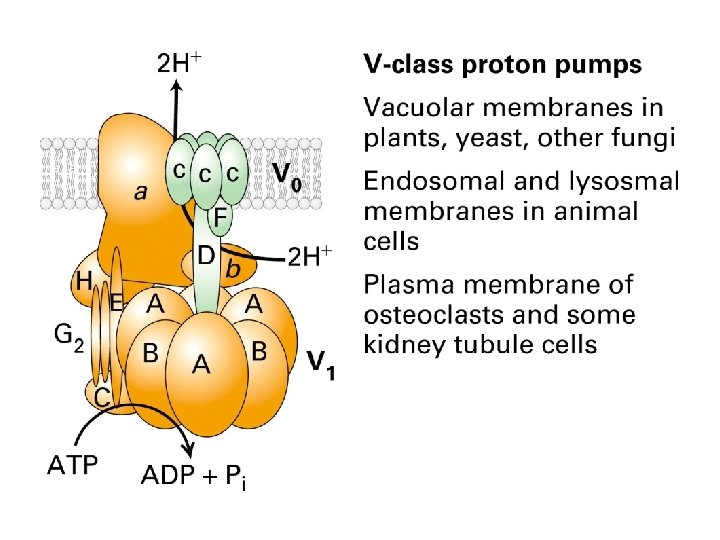
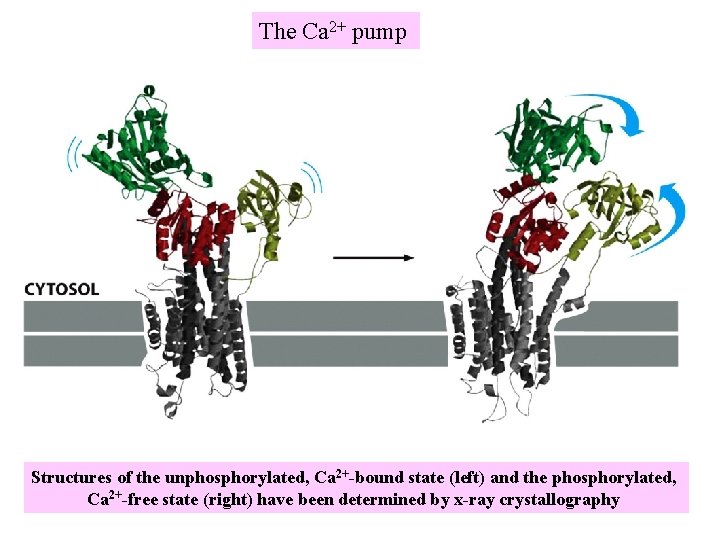
The Ca 2+ pump Structures of the unphosphorylated, Ca 2+-bound state (left) and the phosphorylated, Ca 2+-free state (right) have been determined by x-ray crystallography
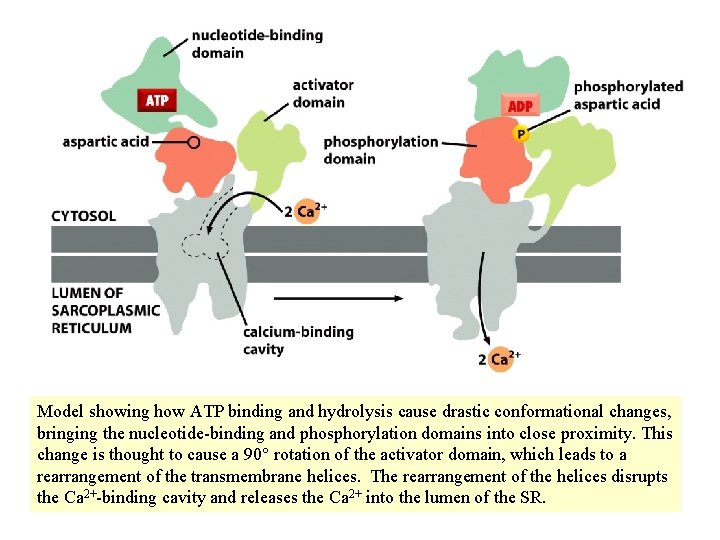
Model showing how ATP binding and hydrolysis cause drastic conformational changes, bringing the nucleotide-binding and phosphorylation domains into close proximity. This change is thought to cause a 90° rotation of the activator domain, which leads to a rearrangement of the transmembrane helices. The rearrangement of the helices disrupts the Ca 2+-binding cavity and releases the Ca 2+ into the lumen of the SR.
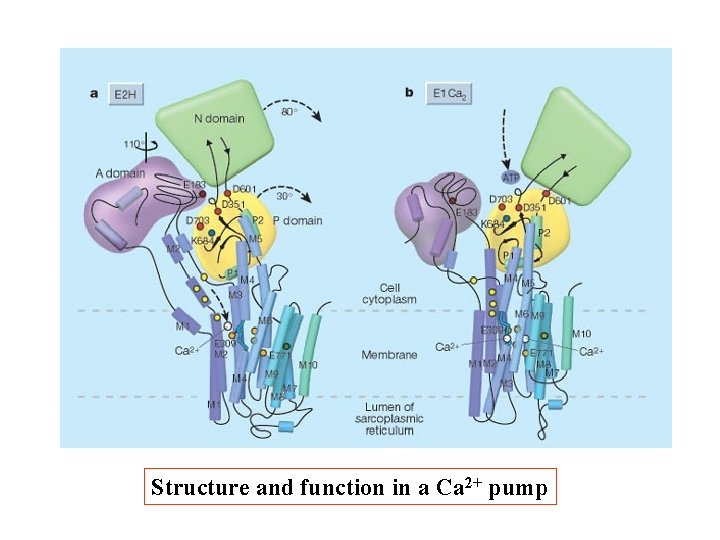
Structure and function in a Ca 2+ pump
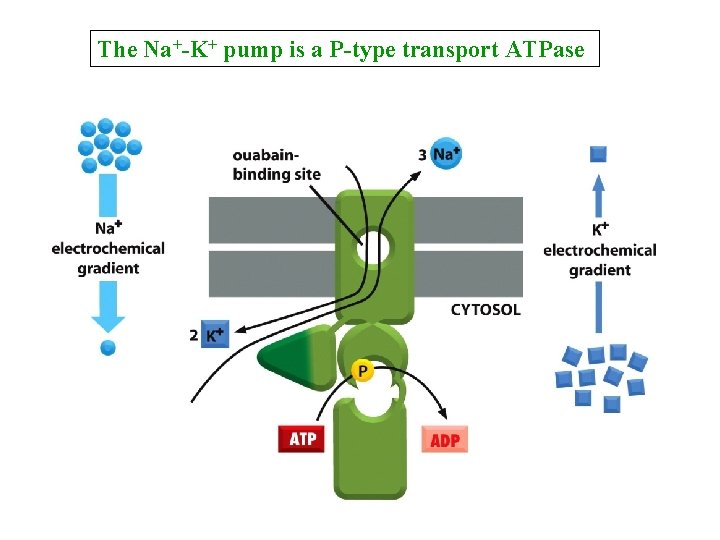
The Na+-K+ pump is a P-type transport ATPase
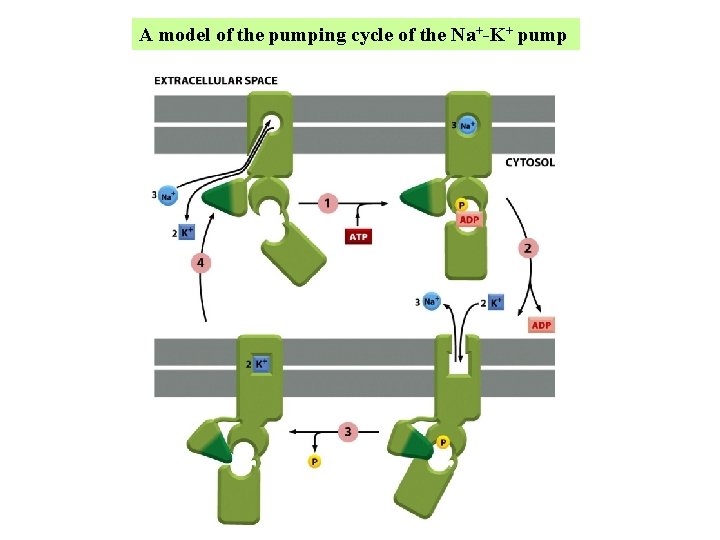
A model of the pumping cycle of the Na+-K+ pump
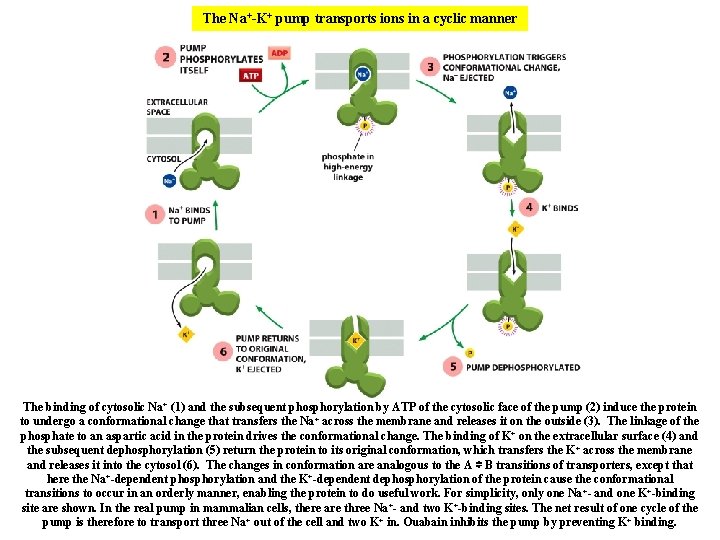
The Na+-K+ pump transports ions in a cyclic manner The binding of cytosolic Na+ (1) and the subsequent phosphorylation by ATP of the cytosolic face of the pump (2) induce the protein to undergo a conformational change that transfers the Na+ across the membrane and releases it on the outside (3). The linkage of the phosphate to an aspartic acid in the protein drives the conformational change. The binding of K+ on the extracellular surface (4) and the subsequent dephosphorylation (5) return the protein to its original conformation, which transfers the K+ across the membrane and releases it into the cytosol (6). The changes in conformation are analogous to the A ⇌ B transitions of transporters, except that here the Na+-dependent phosphorylation and the K+-dependent dephosphorylation of the protein cause the conformational transitions to occur in an orderly manner, enabling the protein to do useful work. For simplicity, only one Na +- and one K+-binding site are shown. In the real pump in mammalian cells, there are three Na +- and two K+-binding sites. The net result of one cycle of the pump is therefore to transport three Na+ out of the cell and two K+ in. Ouabain inhibits the pump by preventing K+ binding.
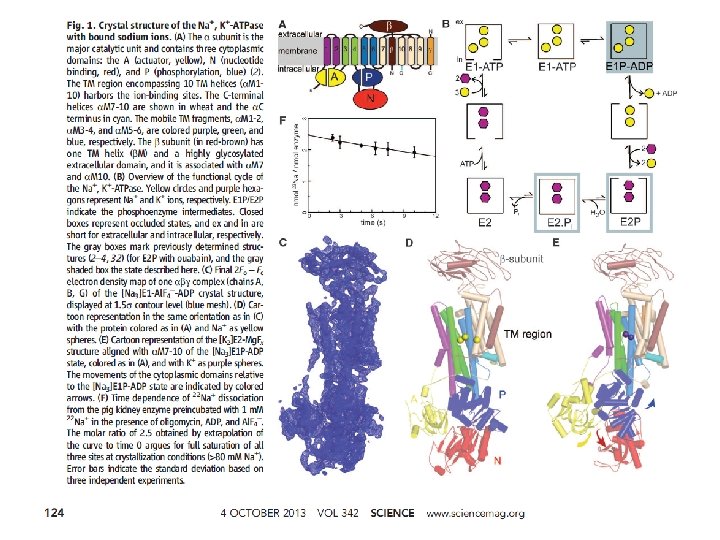
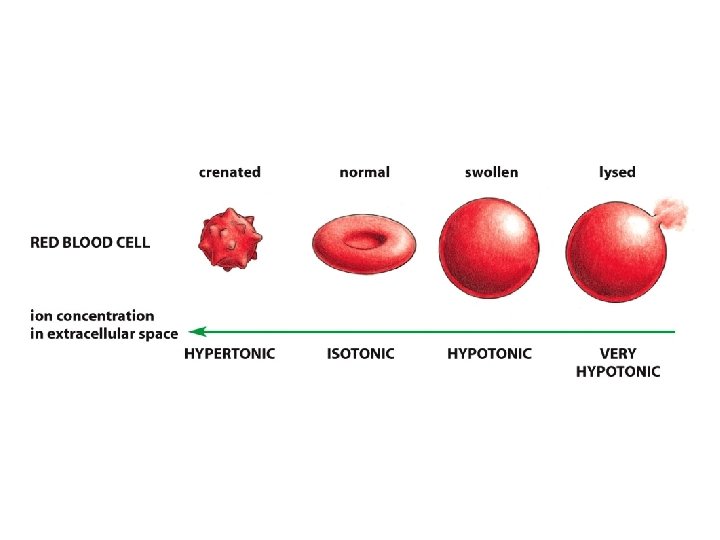
The Na+ - K+ pump is required to maintain osmotic balance and stabilize cell volume
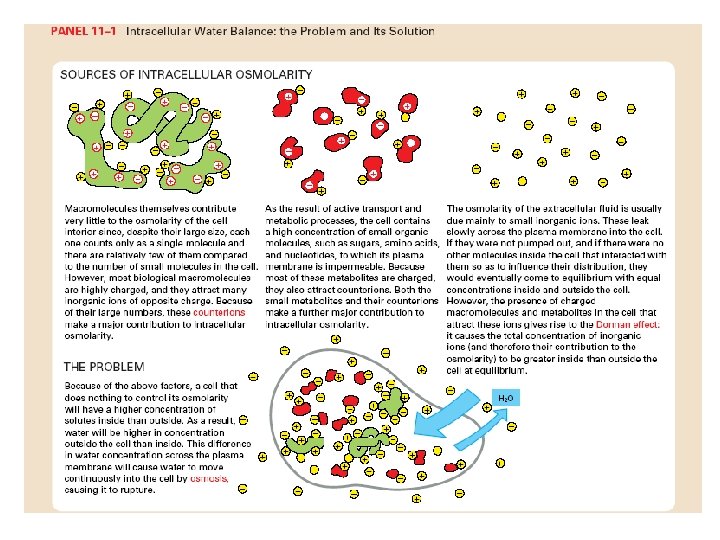
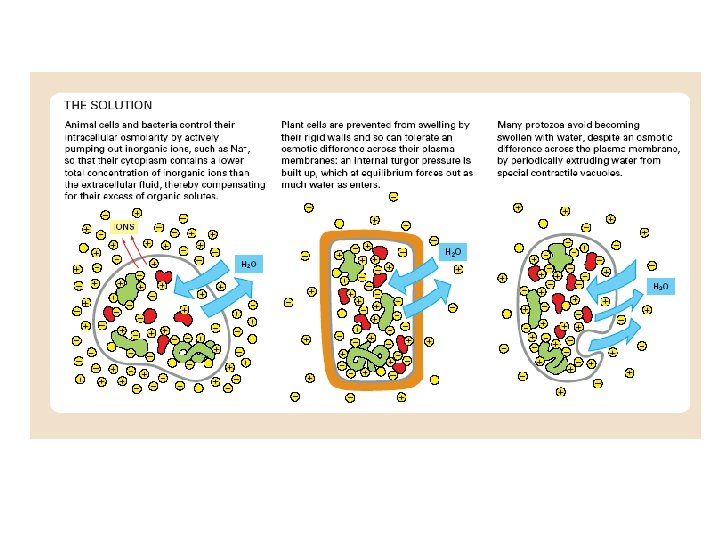

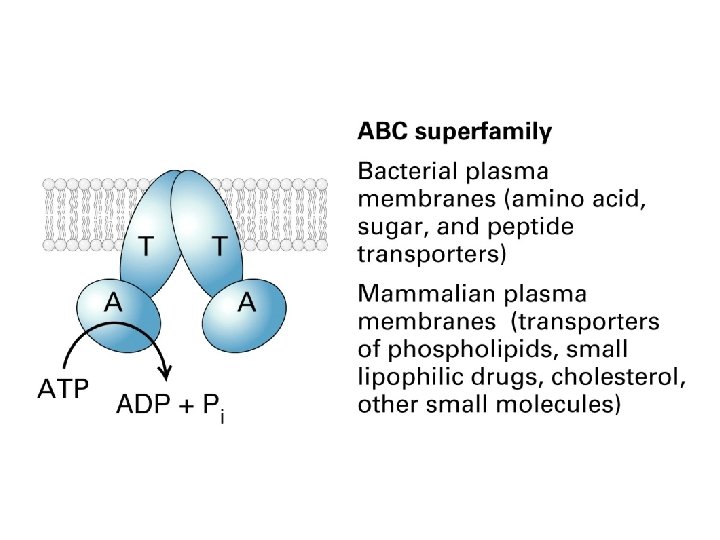
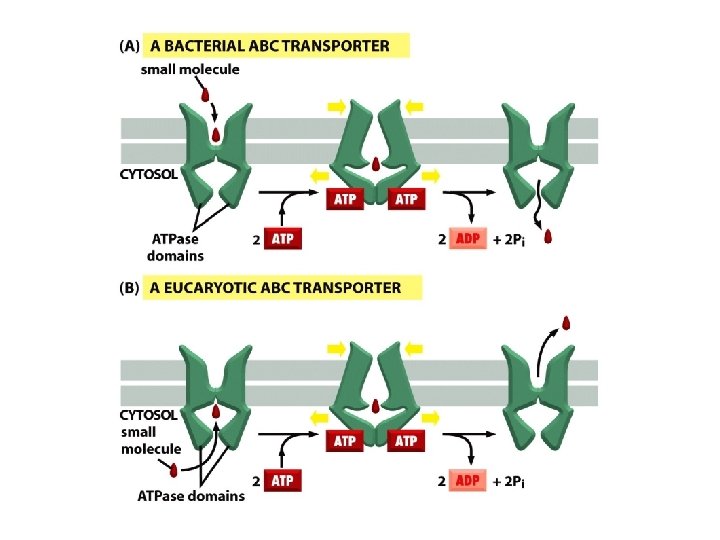
ABC transporters
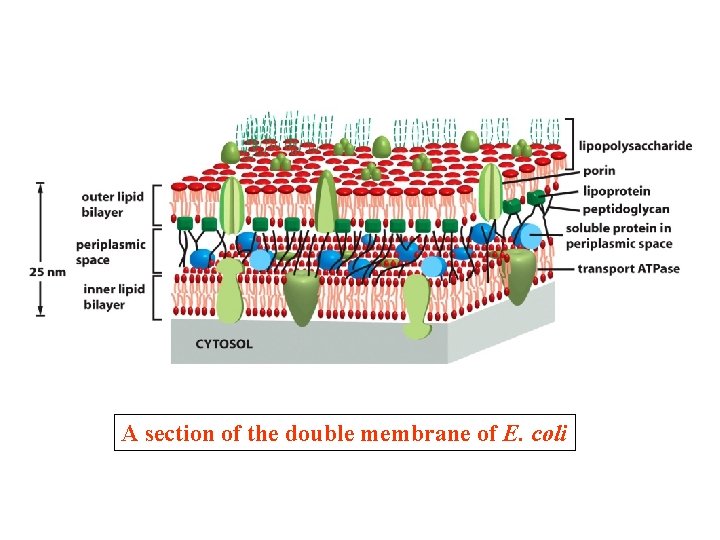
A section of the double membrane of E. coli
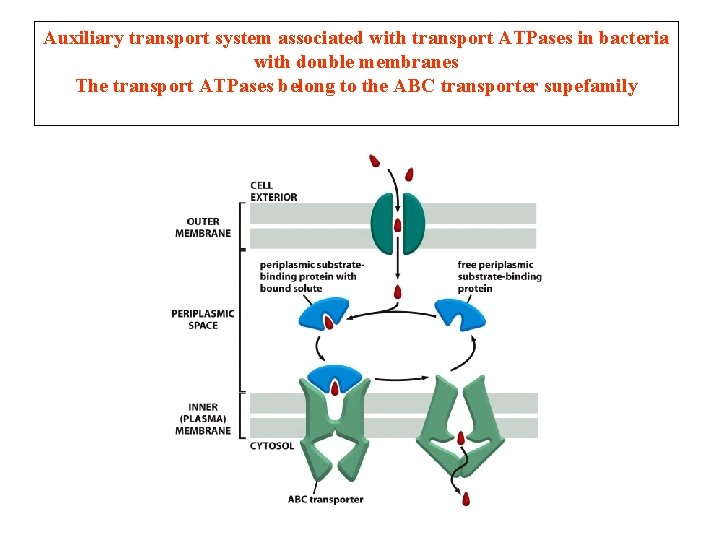
Auxiliary transport system associated with transport ATPases in bacteria with double membranes The transport ATPases belong to the ABC transporter supefamily
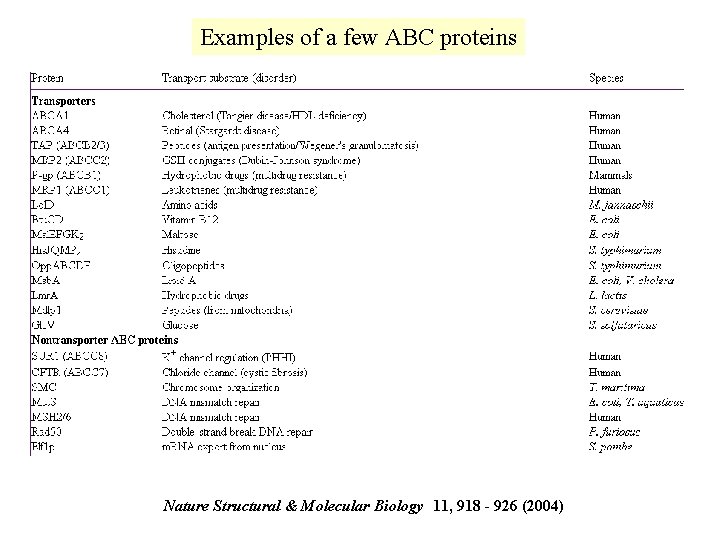
Examples of a few ABC proteins Nature Structural & Molecular Biology 11, 918 - 926 (2004)
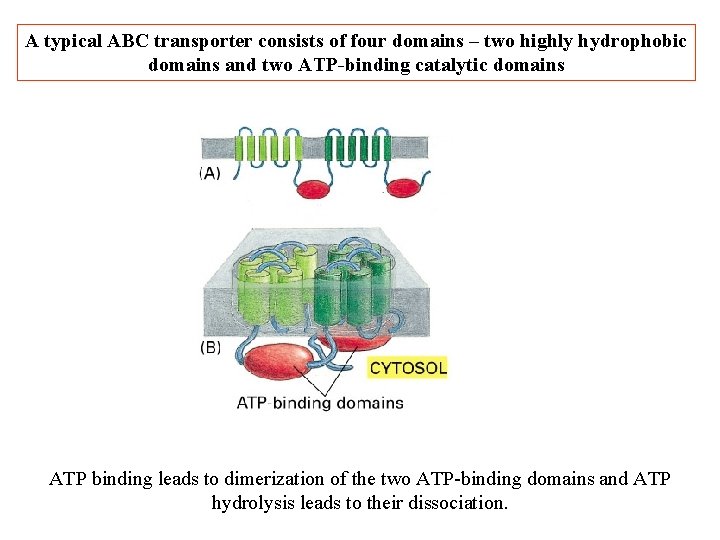
A typical ABC transporter consists of four domains – two highly hydrophobic domains and two ATP-binding catalytic domains ATP binding leads to dimerization of the two ATP-binding domains and ATP hydrolysis leads to their dissociation.
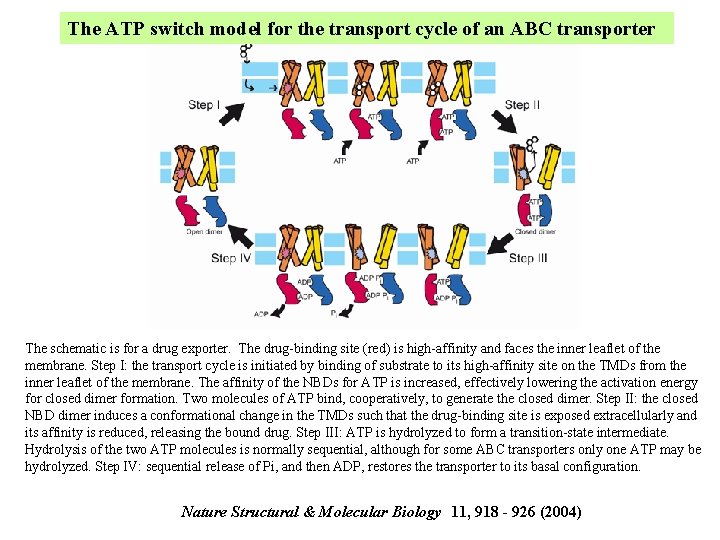
The ATP switch model for the transport cycle of an ABC transporter The schematic is for a drug exporter. The drug-binding site (red) is high-affinity and faces the inner leaflet of the membrane. Step I: the transport cycle is initiated by binding of substrate to its high-affinity site on the TMDs from the inner leaflet of the membrane. The affinity of the NBDs for ATP is increased, effectively lowering the activation energy for closed dimer formation. Two molecules of ATP bind, cooperatively, to generate the closed dimer. Step II: the closed NBD dimer induces a conformational change in the TMDs such that the drug-binding site is exposed extracellularly and its affinity is reduced, releasing the bound drug. Step III: ATP is hydrolyzed to form a transition-state intermediate. Hydrolysis of the two ATP molecules is normally sequential, although for some ABC transporters only one ATP may be hydrolyzed. Step IV: sequential release of Pi, and then ADP, restores the transporter to its basal configuration. Nature Structural & Molecular Biology 11, 918 - 926 (2004)
Ion channels Can transport up to 100 million ions per second, a rate 105 times greater than that mediated by a carrier protein

Among their many functions, ion channels control the pace of the heart, regulate the secretion of hormones into the bloodstream, and generate the electrical impulses underlying information transfer in the nervous system.
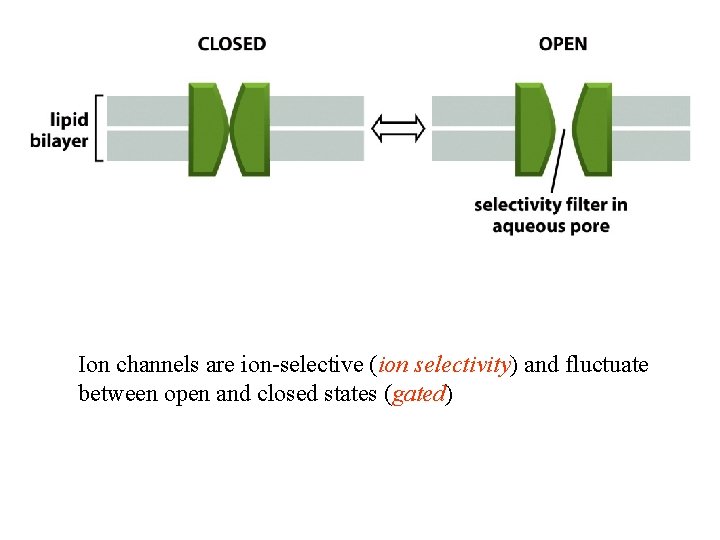
Ion channels are ion-selective (ion selectivity) and fluctuate between open and closed states (gated)

Ion channels, like enzymes, have their specific substrates: potassium, sodium, calcium, and chloride channels permit only their namesake ions to diffuse through their pores. The ability of channels to discriminate among ions is called ion selectivity.

The atomic radius of K+ is 1. 33 Å and that of Na+ is 0. 95 Å With only this difference to work with. K+ channels manage to select for the K+ ion over the Na+ ion by a factor of more than 1000.

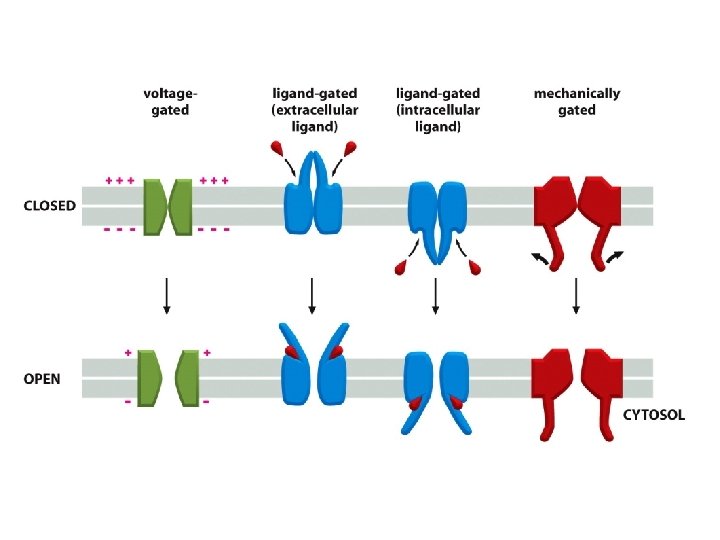
Ion Channel Gating Ion channel gating refers to opening and closing of the ion conduction pore in response to a specific stimulus. Certain channels open when ligands bind (ligand-gated channels); others open in response to membrane voltage (voltage-gated channels); a few open in response to mechanical stress (mechanically gated channels)
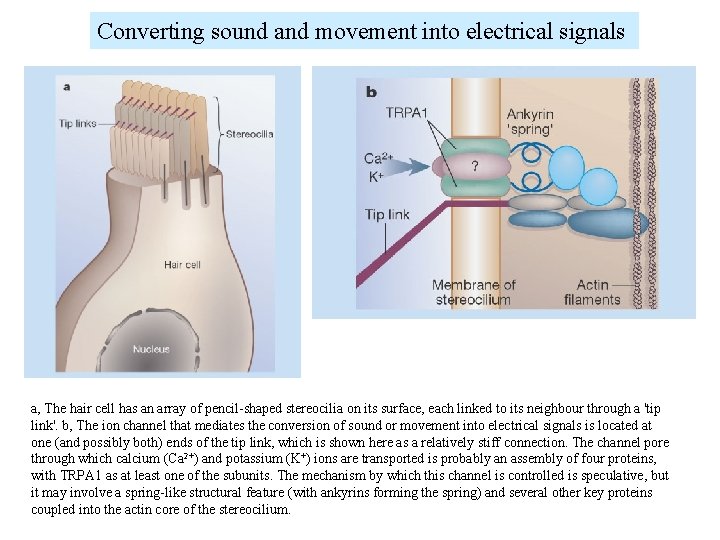
Converting sound and movement into electrical signals a, The hair cell has an array of pencil-shaped stereocilia on its surface, each linked to its neighbour through a 'tip link'. b, The ion channel that mediates the conversion of sound or movement into electrical signals is located at one (and possibly both) ends of the tip link, which is shown here as a relatively stiff connection. The channel pore through which calcium (Ca 2+) and potassium (K+) ions are transported is probably an assembly of four proteins, with TRPA 1 as at least one of the subunits. The mechanism by which this channel is controlled is speculative, but it may involve a spring-like structural feature (with ankyrins forming the spring) and several other key proteins coupled into the actin core of the stereocilium.
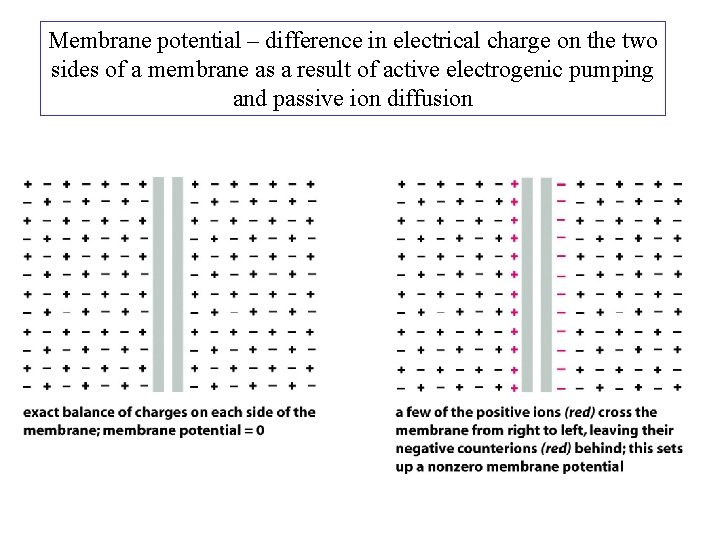
Membrane potential – difference in electrical charge on the two sides of a membrane as a result of active electrogenic pumping and passive ion diffusion
The equilibrium condition, in which there is no net flow of ions across the plasma membrane defines the resting membrane potential The Nernst equation expresses the equilibrium condition quantitatively

The number of ions that move across the plasma membrane to set up the membrane potential is minute and the movements of charge are generally rapid
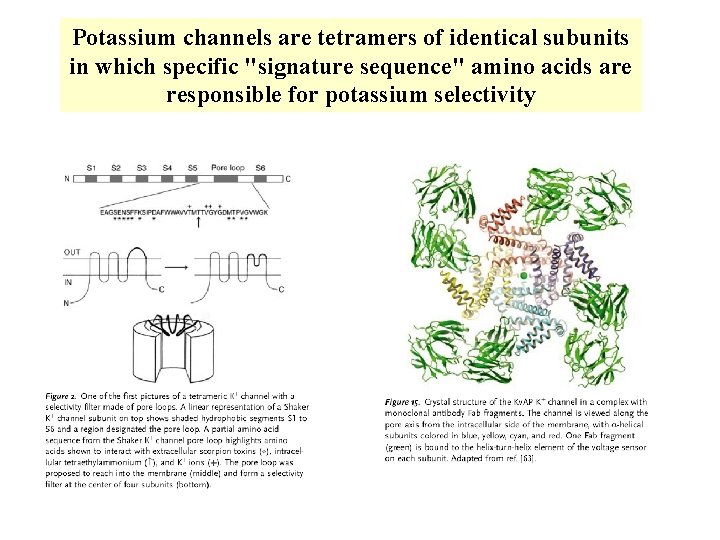
Potassium channels are tetramers of identical subunits in which specific "signature sequence" amino acids are responsible for potassium selectivity
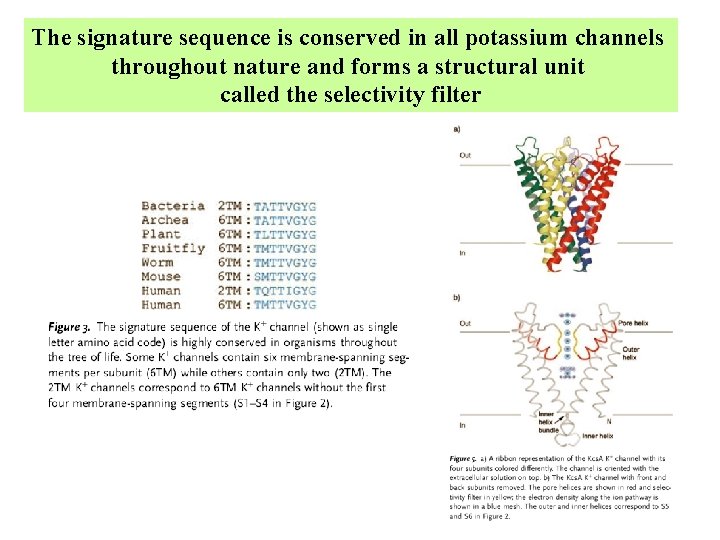
The signature sequence is conserved in all potassium channels throughout nature and forms a structural unit called the selectivity filter
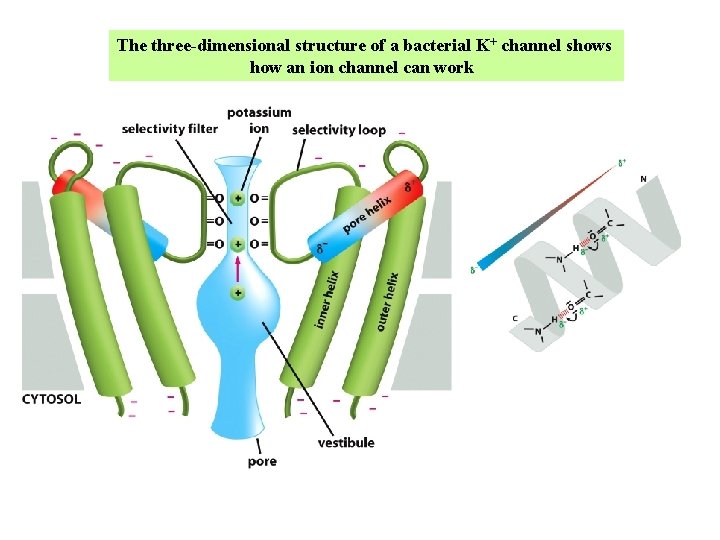
The three-dimensional structure of a bacterial K+ channel shows how an ion channel can work
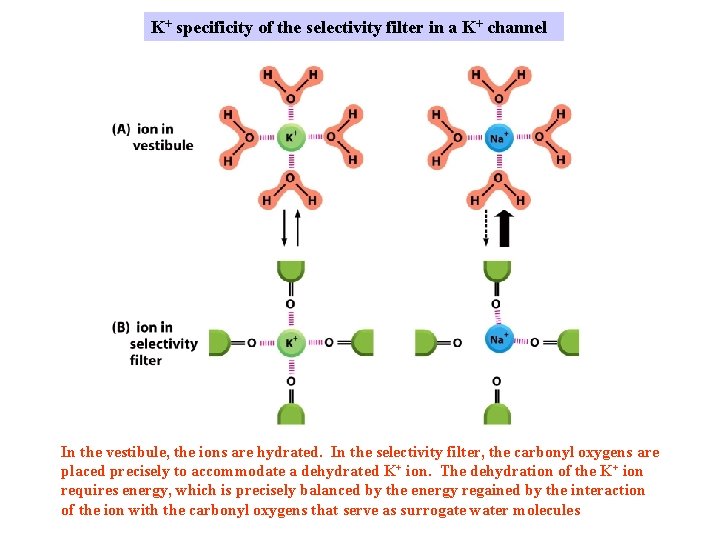
K+ specificity of the selectivity filter in a K+ channel In the vestibule, the ions are hydrated. In the selectivity filter, the carbonyl oxygens are placed precisely to accommodate a dehydrated K+ ion. The dehydration of the K+ ion requires energy, which is precisely balanced by the energy regained by the interaction of the ion with the carbonyl oxygens that serve as surrogate water molecules


EACH OF THE binding sites closely mimics potassium ions' octahedral hydration shell, thereby minimizing the energy required to strip off their water coats. Because of their smaller size, sodium ions don't fit in these binding sites as snugly and thus find the energetic cost of trading their water coat for a spot in the selectivity filter too high.
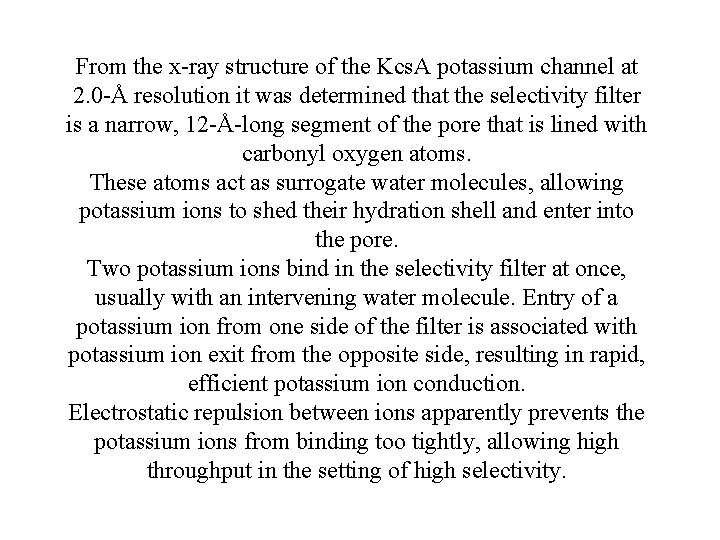
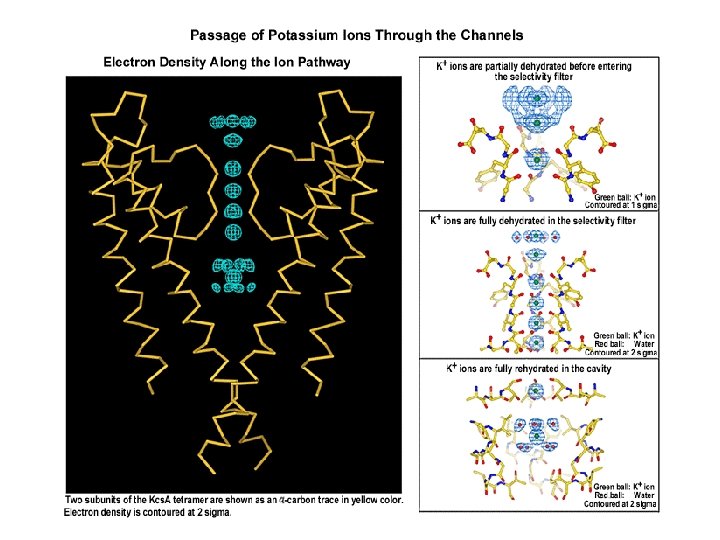
From the x-ray structure of the Kcs. A potassium channel at 2. 0 -Å resolution it was determined that the selectivity filter is a narrow, 12 -Å-long segment of the pore that is lined with carbonyl oxygen atoms. These atoms act as surrogate water molecules, allowing potassium ions to shed their hydration shell and enter into the pore. Two potassium ions bind in the selectivity filter at once, usually with an intervening water molecule. Entry of a potassium ion from one side of the filter is associated with potassium ion exit from the opposite side, resulting in rapid, efficient potassium ion conduction. Electrostatic repulsion between ions apparently prevents the potassium ions from binding too tightly, allowing high throughput in the setting of high selectivity.

A marvelous electrostatic feature of the potassium channel is a cavity of water half way across the membrane with specifically oriented a helices. This design reduces the dielectric barrier, that is, the energetic cost of moving an ion from the high-dielectric environment of water to the low-dielectric membrane, and thereby supports a high ion throughput.
Ion selectivity K+ channels specifically conduct K+ ions because the selectivity filter contains multiple binding sites that mimic a hydrated K+ ion’s hydration shell. Potassium channels achieve high conduction rates by exploiting electrostatic repulsion between closely spaced ions and by coupling the conformation of the selectivity filter to ion binding within the filter

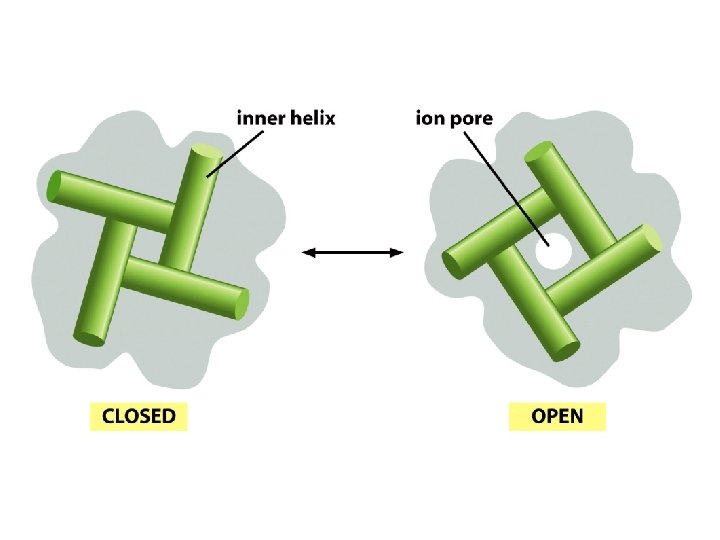
Gating Large conformational changes within the membrane underlie pore opening in K+ channels. Inner helices obstruct the pore in the closed state and expand its intracellular diameter in the opened state. Different gating domains – ligand binding and voltage sensingappear to bring about a similar conformational change in the pore of K+ channels

Voltage sensing Conformational changes underlying voltage sensing in Kv. AP are large and involve movements of arginine residues through the membrane, near the protein lipid interface
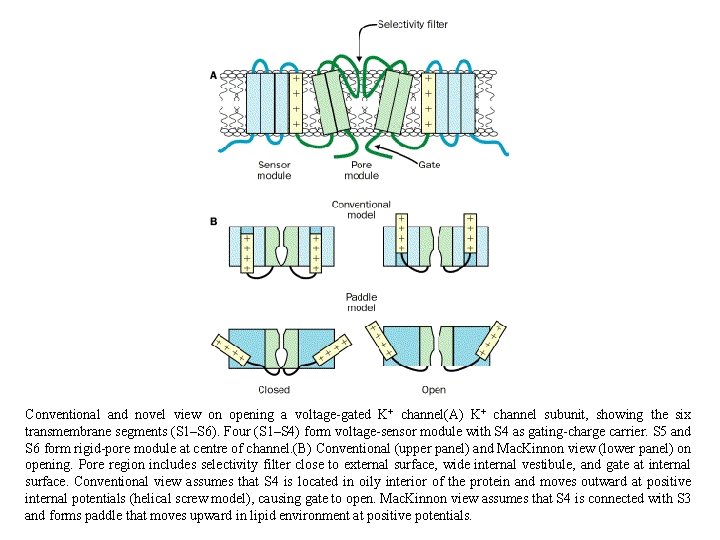
Conventional and novel view on opening a voltage-gated K+ channel(A) K+ channel subunit, showing the six transmembrane segments (S 1–S 6). Four (S 1–S 4) form voltage-sensor module with S 4 as gating-charge carrier. S 5 and S 6 form rigid-pore module at centre of channel. (B) Conventional (upper panel) and Mac. Kinnon view (lower panel) on opening. Pore region includes selectivity filter close to external surface, wide internal vestibule, and gate at internal surface. Conventional view assumes that S 4 is located in oily interior of the protein and moves outward at positive internal potentials (helical screw model), causing gate to open. Mac. Kinnon view assumes that S 4 is connected with S 3 and forms paddle that moves upward in lipid environment at positive potentials.
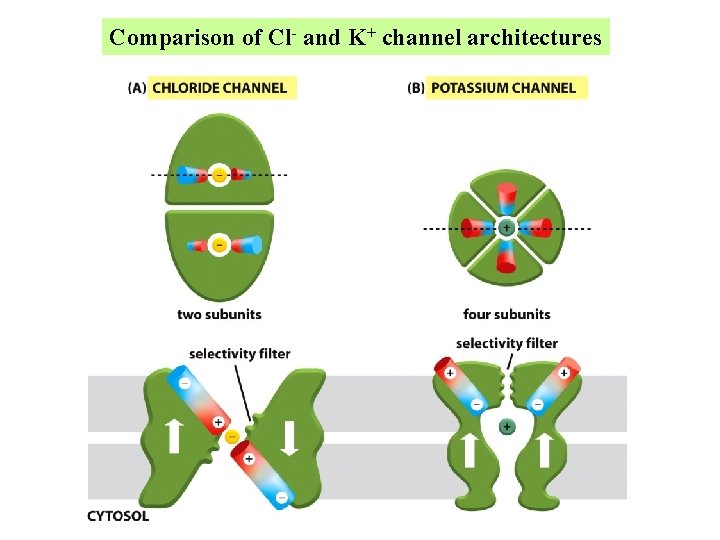
Comparison of Cl- and K+ channel architectures

Aquaporins are permeable to water but impermeable to ions
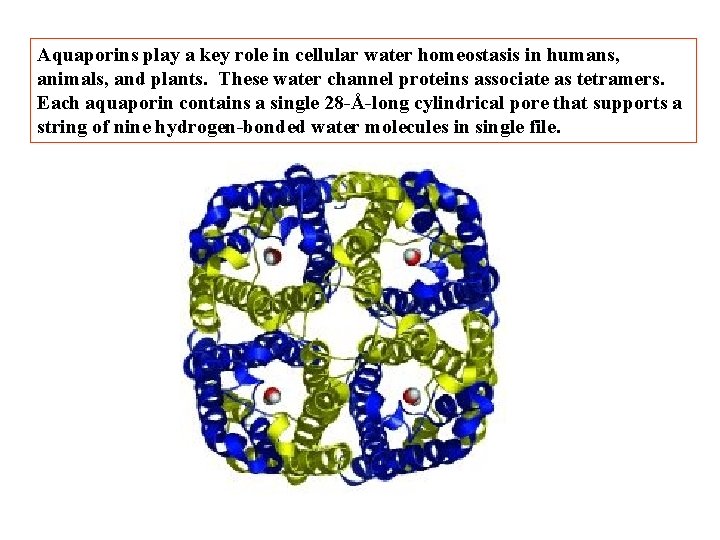
Aquaporins play a key role in cellular water homeostasis in humans, animals, and plants. These water channel proteins associate as tetramers. Each aquaporin contains a single 28 -Å-long cylindrical pore that supports a string of nine hydrogen-bonded water molecules in single file.
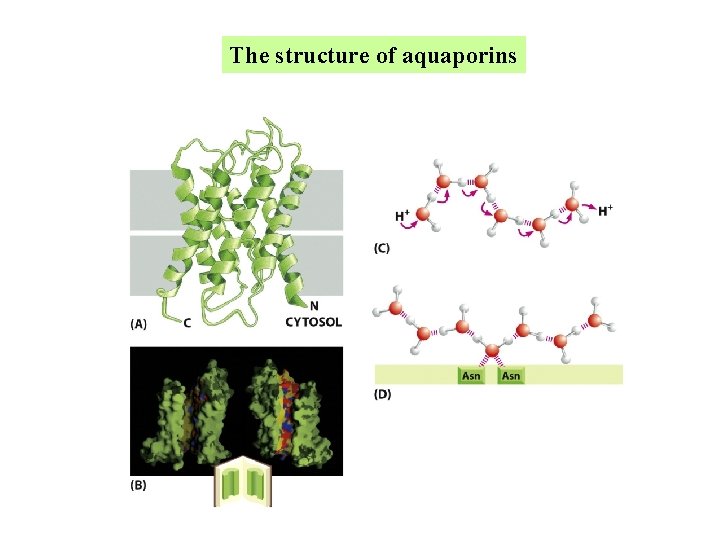
The structure of aquaporins
Animations of the movement of water through aquaporins:
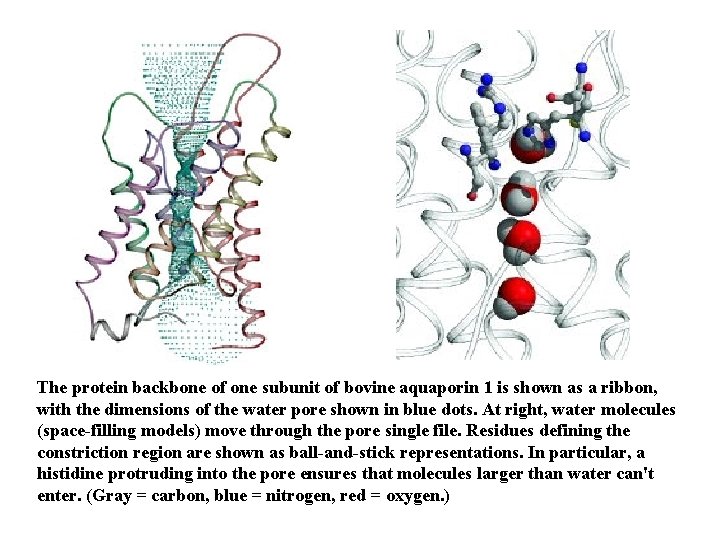
The protein backbone of one subunit of bovine aquaporin 1 is shown as a ribbon, with the dimensions of the water pore shown in blue dots. At right, water molecules (space-filling models) move through the pore single file. Residues defining the constriction region are shown as ball-and-stick representations. In particular, a histidine protruding into the pore ensures that molecules larger than water can't enter. (Gray = carbon, blue = nitrogen, red = oxygen. )

High-resolution structures reveal the inner workings of aquaporins in atomic detail Water molecules travel single file through a narrow pore that connects water-filled intracellular and extracellular vestibules. This 28 -Å-long pore is lined with a row of eight carbonyl oxygen atoms that can accept hydrogen bonds from the queue of water molecules, "ensuring that every water molecule is precisely oriented throughout the channel. " But because the rest of the pore is lined with mostly hydrophobic amino acids, "waterpore interactions are kept to a minimum, " allowing water to rush through each pore at a rate of 3 x 109 molecules per second.

FOR A SHORT STRETCH, the pore narrows to just wide enough for a water molecule to pass, preventing larger molecules from entering. And ions--which would have to abandon their bulky water coats to enter--don't do so because the pore isn't a good substitute. In addition, a conserved, positively charged arginine residue in this region prevents cations, including protonated water (H 3 O+), from entering. Nature has gone to great effort to prevent protons from passing through, too. Previous structural work had suggested that a pair of asparagine residues as well as the dipole moments of short a-helices would force water molecules to flip near the pore's center. This forced reorientation breaks the continuous chain of hydrogen-bonded waters and prevents protons from "hopping" along the hydrogen-bond network.
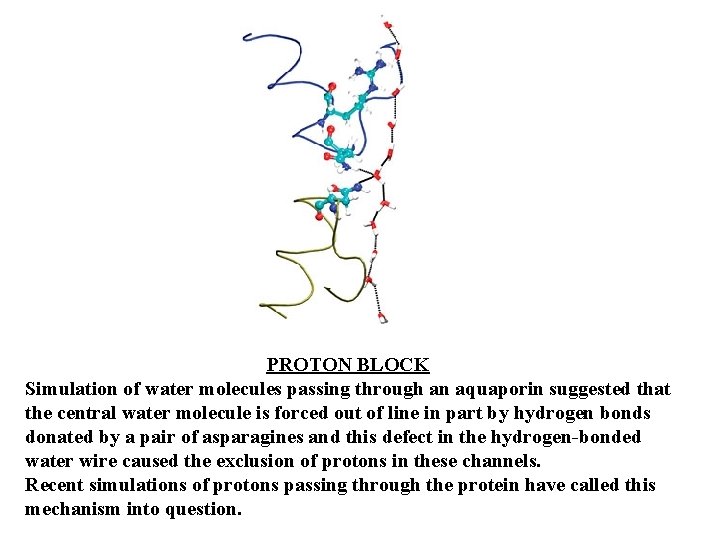
PROTON BLOCK Simulation of water molecules passing through an aquaporin suggested that the central water molecule is forced out of line in part by hydrogen bonds donated by a pair of asparagines and this defect in the hydrogen-bonded water wire caused the exclusion of protons in these channels. Recent simulations of protons passing through the protein have called this mechanism into question.

Normally, continuous, single-file columns of water like these conduct protons. Protons can easily hop along a similar single-file water column in the water pore of gramicidin A. But unlike gramicidin A, an antibiotic peptide that kills bacteria by forming pores in their cell membranes, aquaporins must not leak protons. Doing so would disturb the delicate balance of charge across cell membranes that underlies cellular function.

So how do aquaporins manage to do what gramicidin A can't? The water column in aquaporins does have some distinguishing features. Near the center of the aquaporin pore, hydrogen bonds from a pair of asparagine residues (as well as the pull of nearby a-helix dipoles) reorient the central water molecule, preventing it from accepting a proton from nearby water molecules. From structural observations, it was suggested that the forced reorientation of this central water molecule might break the continuous chain of hydrogen-bonded waters and possibly prevent protons from "hopping" along the hydrogen-bonded water "wire. "

The barrier to proton transfer is highest in the part of the aquaporin pore that boasts the highest electrostatic potential: the region centered around the two central asparagines and the partially positively charged a-helix dipoles, known as the NPA region. Recent results indicate that the main barrier to proton transfer is not caused by interruption of the hydrogen-bonded water chain, as had previously been speculated, but rather by an electrostatic field created by the a-helix dipoles in the NPA region. The barrier to proton transfer in aquaporins is estimated to be 6– 7 kcal per mole--about the same as estimates of the barrier to proton translocation across lipid membranes
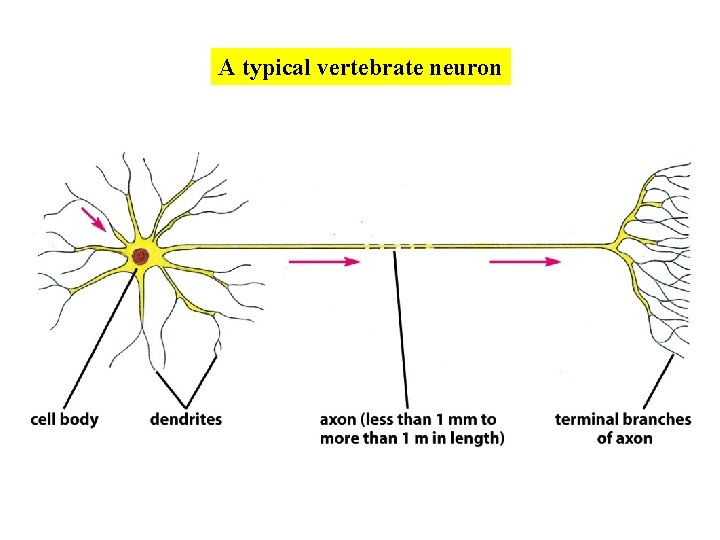
A typical vertebrate neuron

Voltage-gated cation channels generate action potentials in electrically excitable cells - neurons - muscle cells - endocrine cells - egg cells
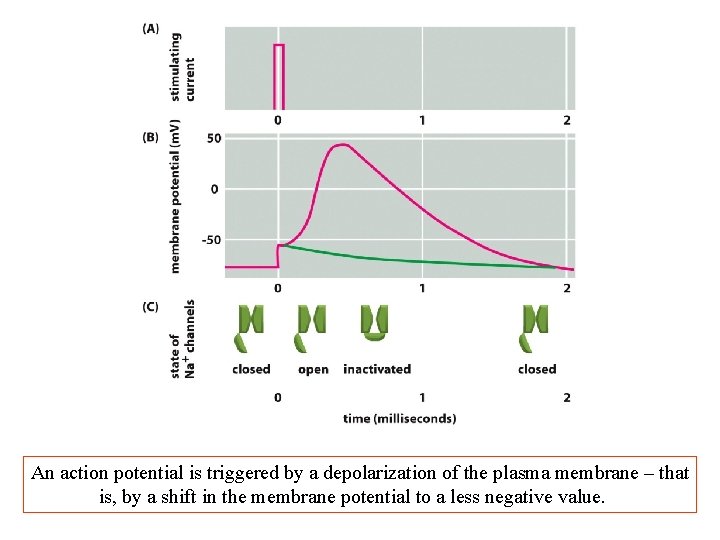
An action potential is triggered by a depolarization of the plasma membrane – that is, by a shift in the membrane potential to a less negative value.
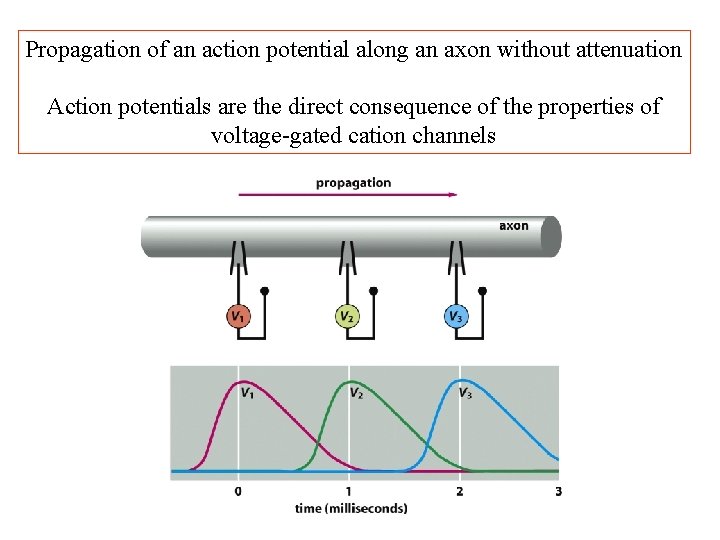
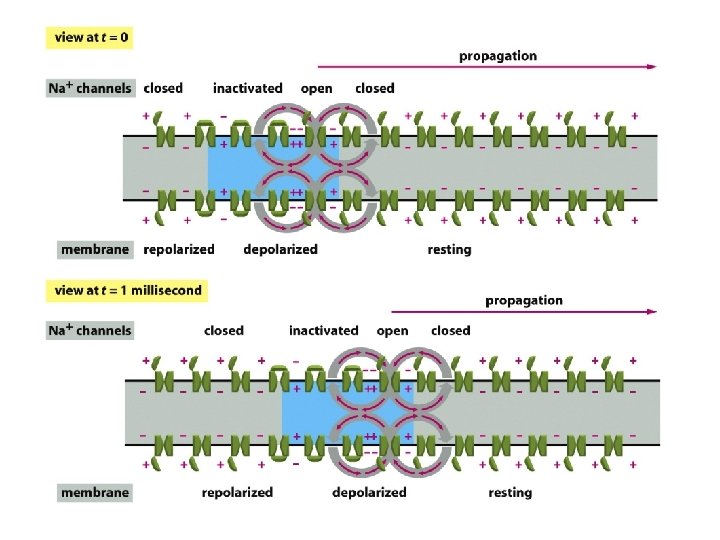
Propagation of an action potential along an axon without attenuation Action potentials are the direct consequence of the properties of voltage-gated cation channels
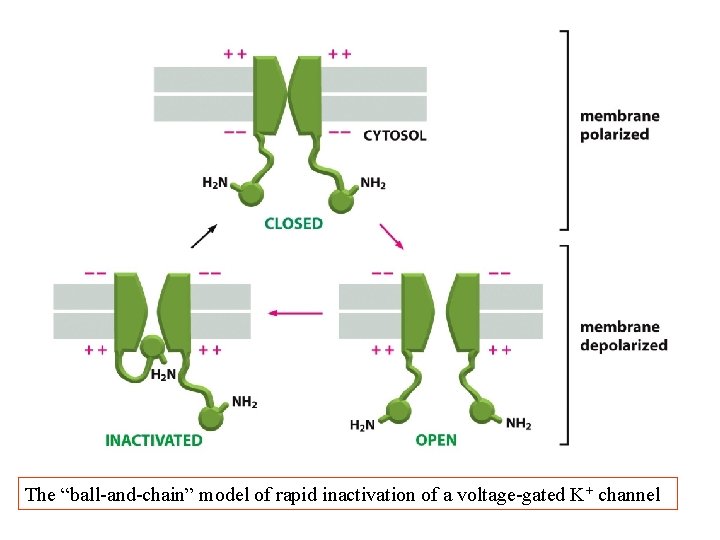
The “ball-and-chain” model of rapid inactivation of a voltage-gated K+ channel
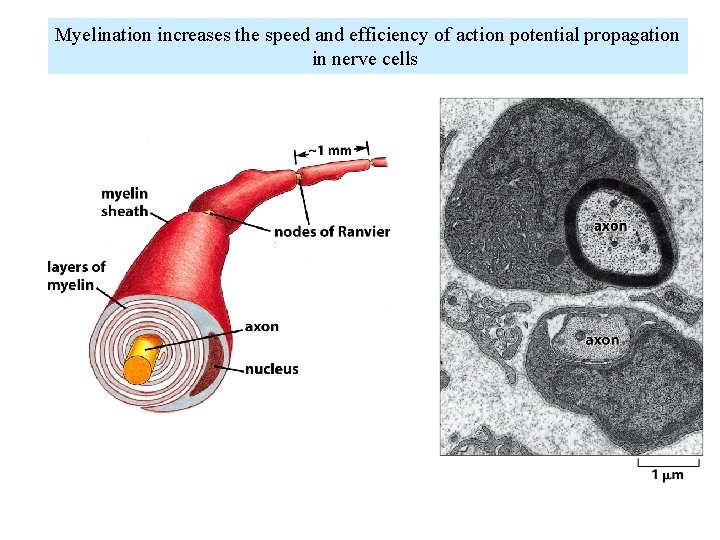
Myelination increases the speed and efficiency of action potential propagation in nerve cells
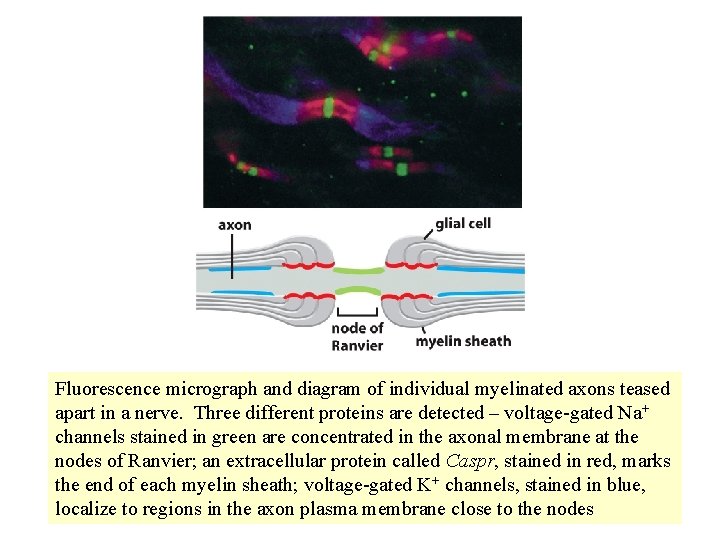
Fluorescence micrograph and diagram of individual myelinated axons teased apart in a nerve. Three different proteins are detected – voltage-gated Na+ channels stained in green are concentrated in the axonal membrane at the nodes of Ranvier; an extracellular protein called Caspr, stained in red, marks the end of each myelin sheath; voltage-gated K+ channels, stained in blue, localize to regions in the axon plasma membrane close to the nodes
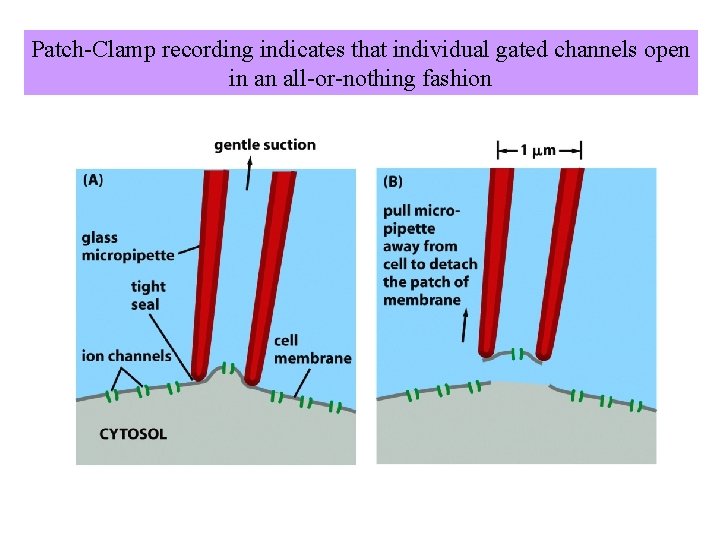
Patch-Clamp recording indicates that individual gated channels open in an all-or-nothing fashion
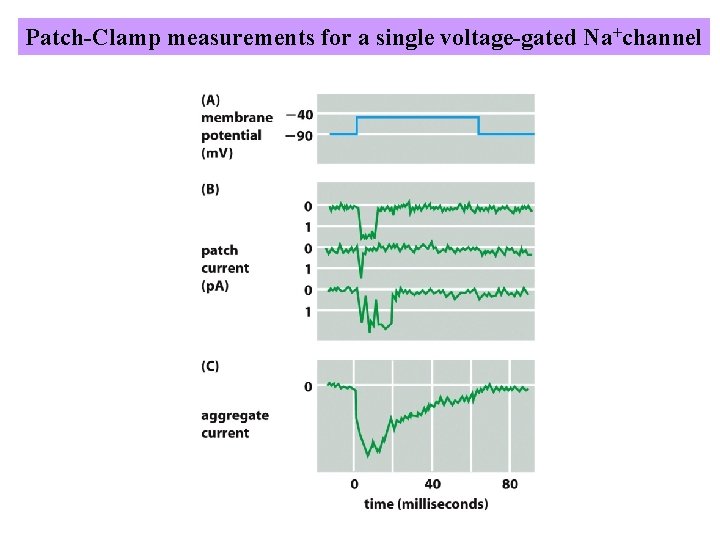
Patch-Clamp measurements for a single voltage-gated Na+channel
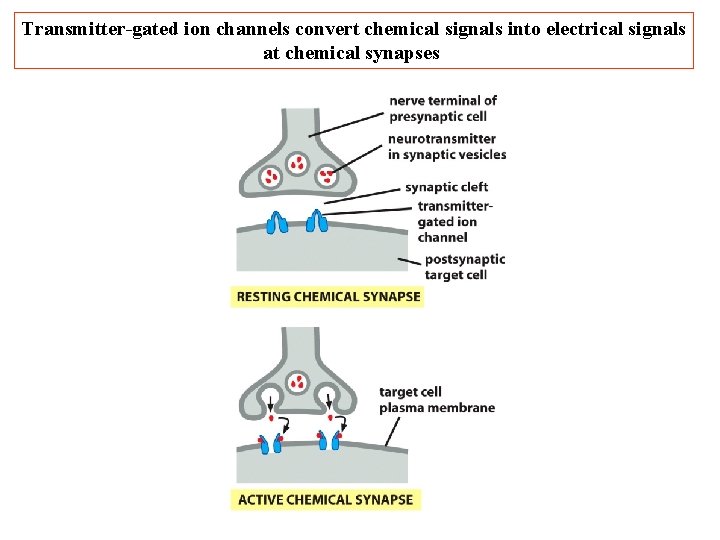
Transmitter-gated ion channels convert chemical signals into electrical signals at chemical synapses
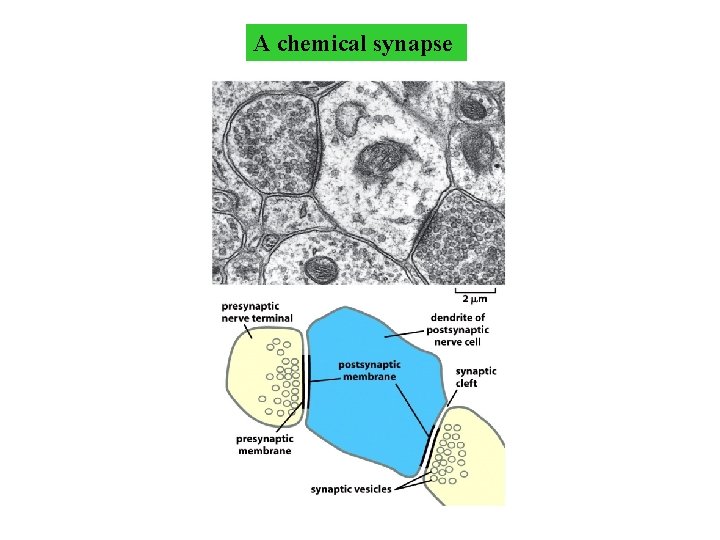
A chemical synapse
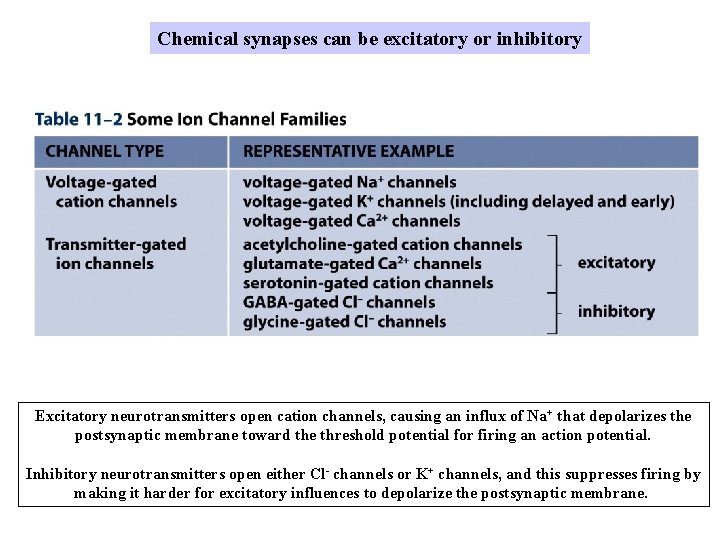
Chemical synapses can be excitatory or inhibitory Excitatory neurotransmitters open cation channels, causing an influx of Na+ that depolarizes the postsynaptic membrane toward the threshold potential for firing an action potential. Inhibitory neurotransmitters open either Cl- channels or K+ channels, and this suppresses firing by making it harder for excitatory influences to depolarize the postsynaptic membrane.
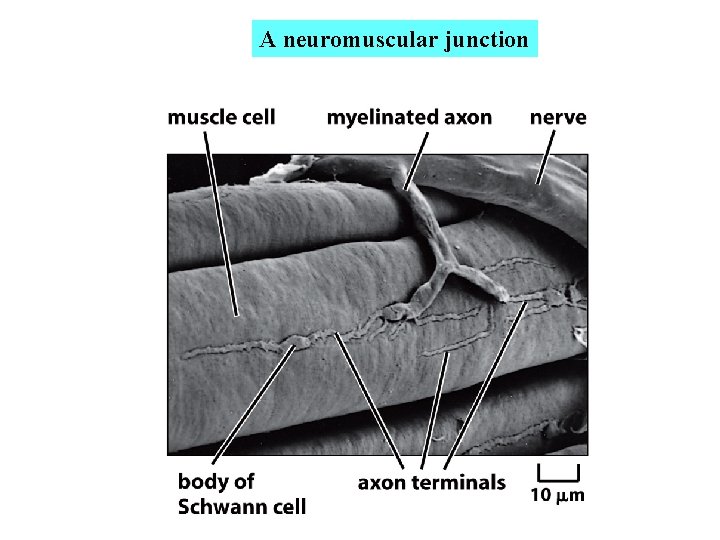
A neuromuscular junction
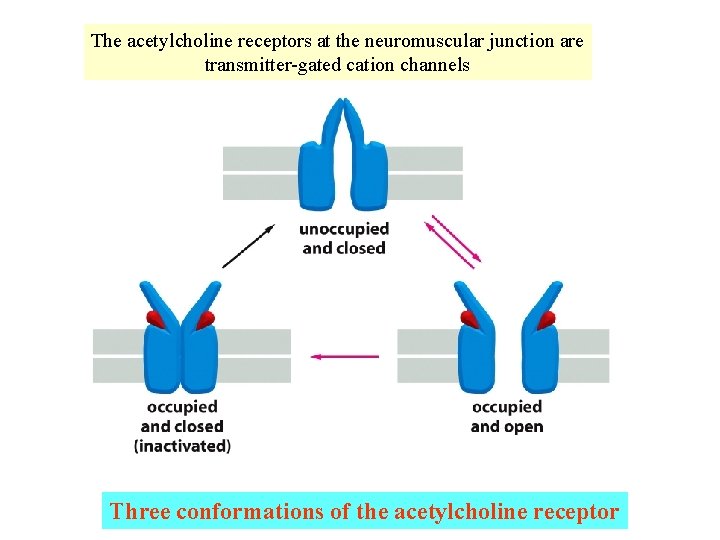
The acetylcholine receptors at the neuromuscular junction are transmitter-gated cation channels Three conformations of the acetylcholine receptor
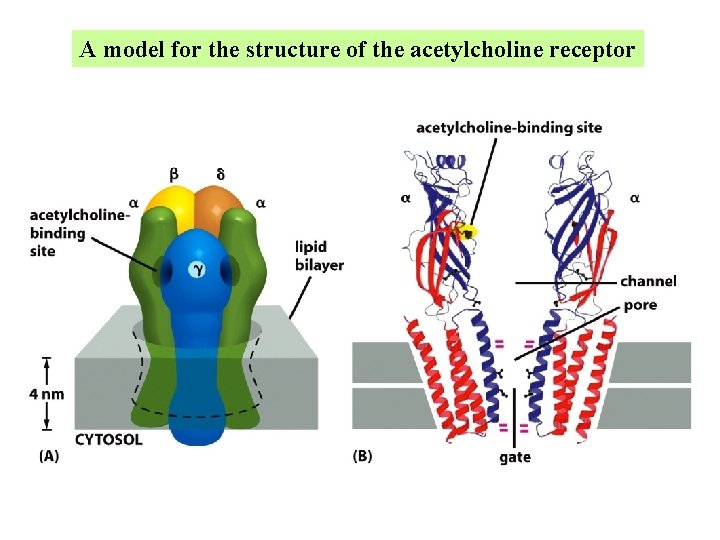
A model for the structure of the acetylcholine receptor
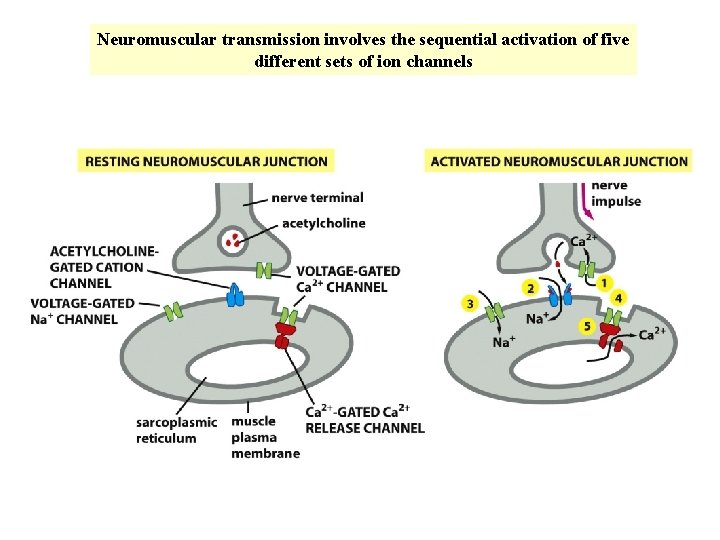
Neuromuscular transmission involves the sequential activation of five different sets of ion channels
- Slides: 101