CHAPTER 10 Regulation of Gene Expression in Bacteria

CHAPTER 10 Regulation of Gene Expression in Bacteria and Their Viruses Copyright 2008 © W H Freeman and Company
CHAPTER OUTLINE 10. 1 Gene regulation 10. 2 Discovery of the lac system: negative control 10. 3 Catabolite repression of the lac operon: positive control 10. 4 Dual positive and negative control: the arabinose operon 10. 5 Metabolic pathways and additional levels of regulation: attenuation 10. 6 Bacteriophage life cycles: more regulators, complex operons 10. 7 Alternative sigma factors regulate large sets of genes
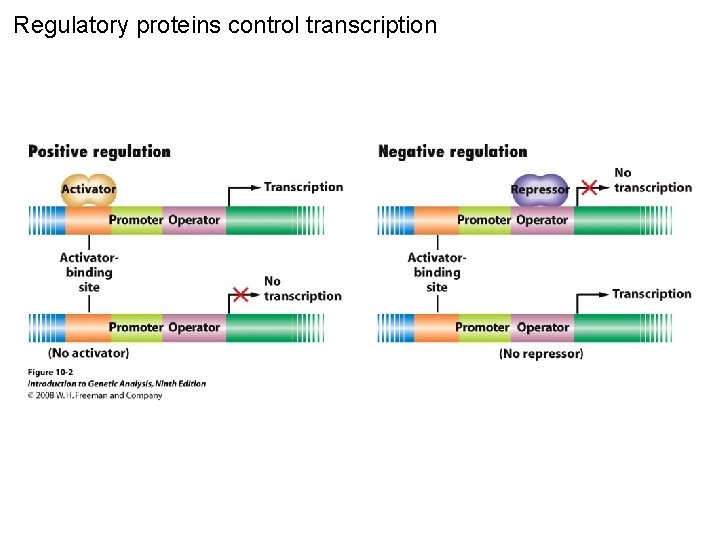
Regulatory proteins control transcription Figure 10 -2
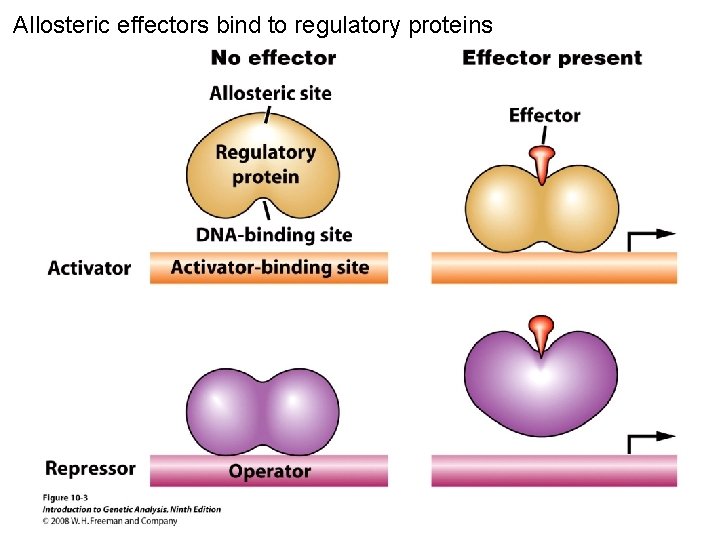
Allosteric effectors bind to regulatory proteins Figure 10 -3
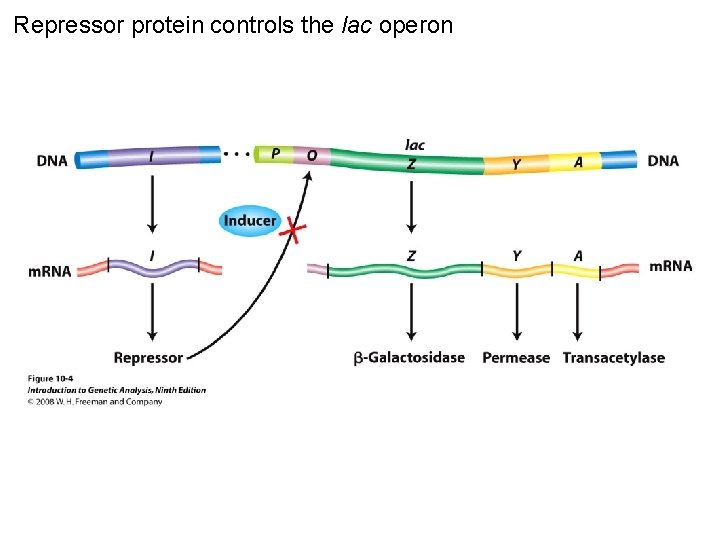
Repressor protein controls the lac operon Figure 10 -4
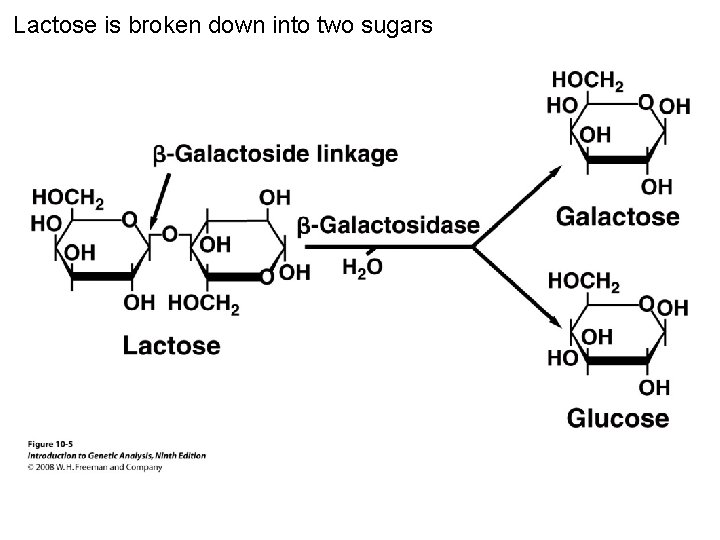
Lactose is broken down into two sugars Figure 10 -5
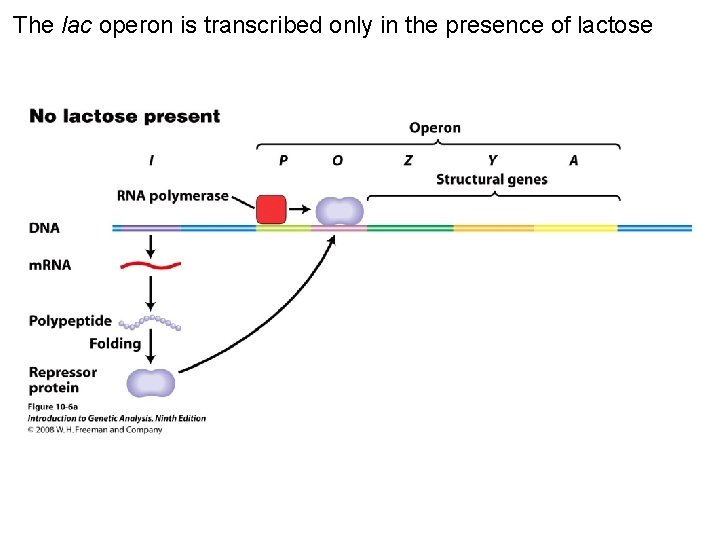
The lac operon is transcribed only in the presence of lactose Figure 10 -6 a
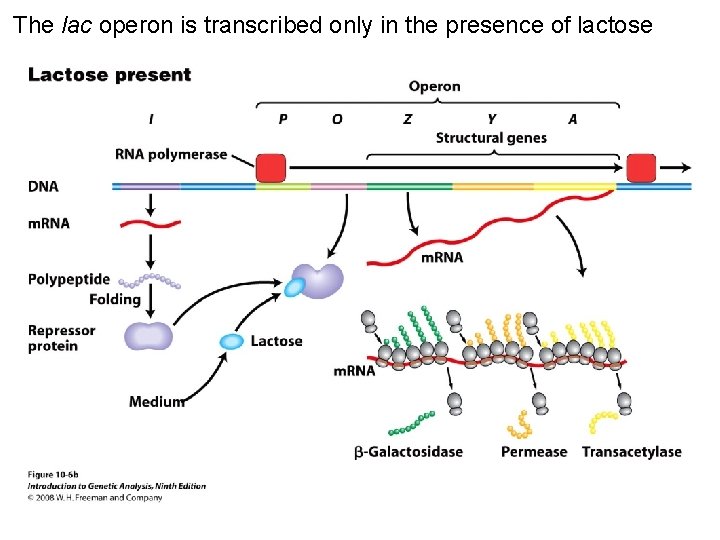
The lac operon is transcribed only in the presence of lactose Figure 10 -6 b
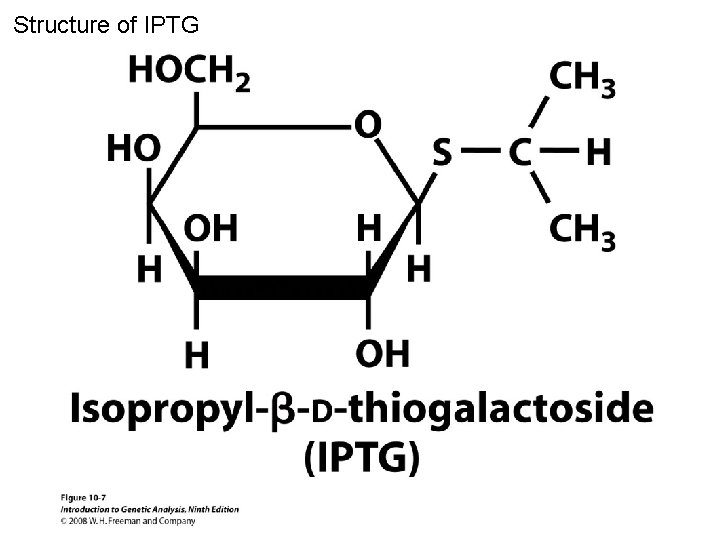
Structure of IPTG Figure 10 -7
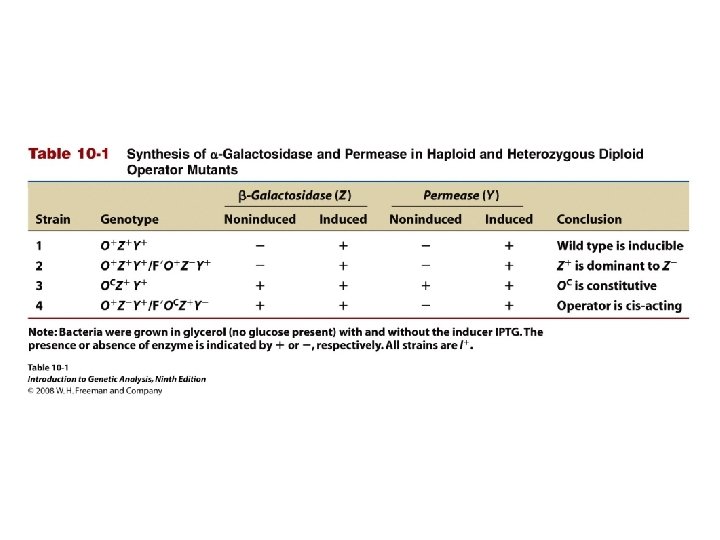
Table 10 -1
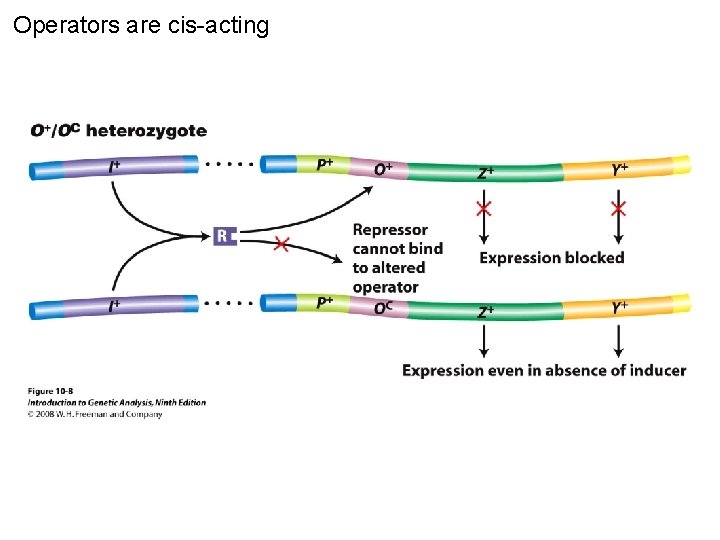
Operators are cis-acting Figure 10 -8
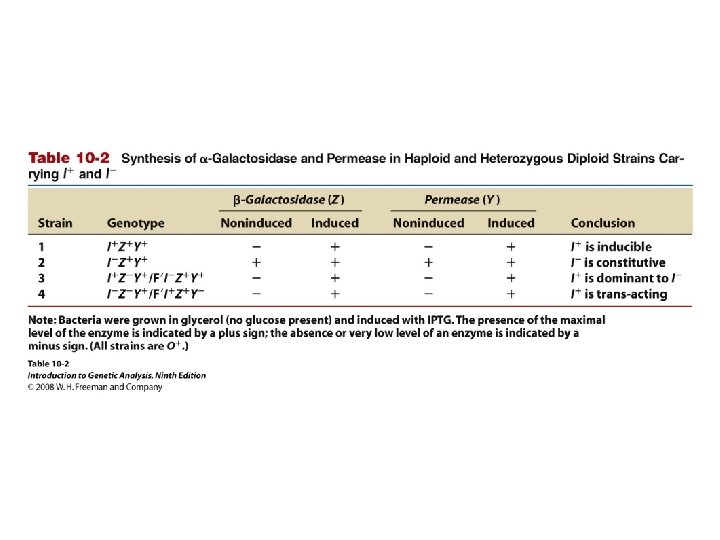
Table 10 -2
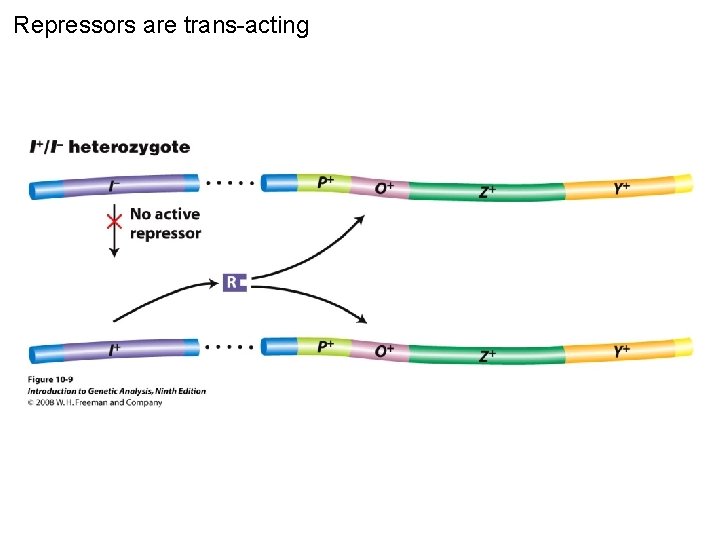
Repressors are trans-acting Figure 10 -9
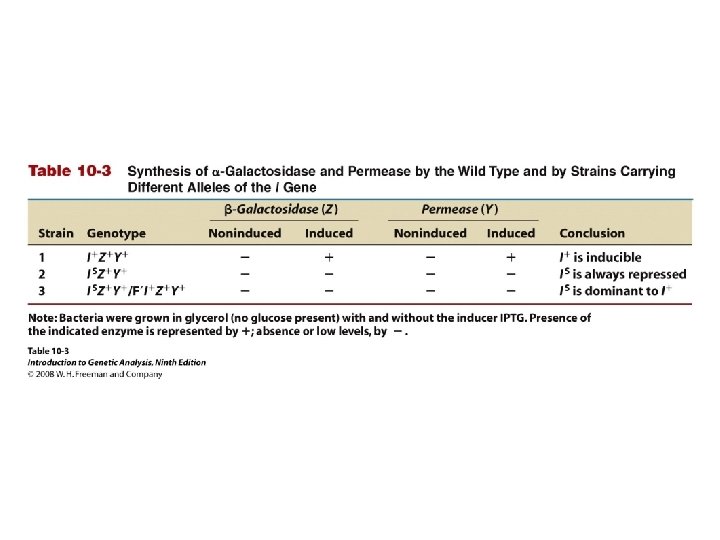
Table 10 -3
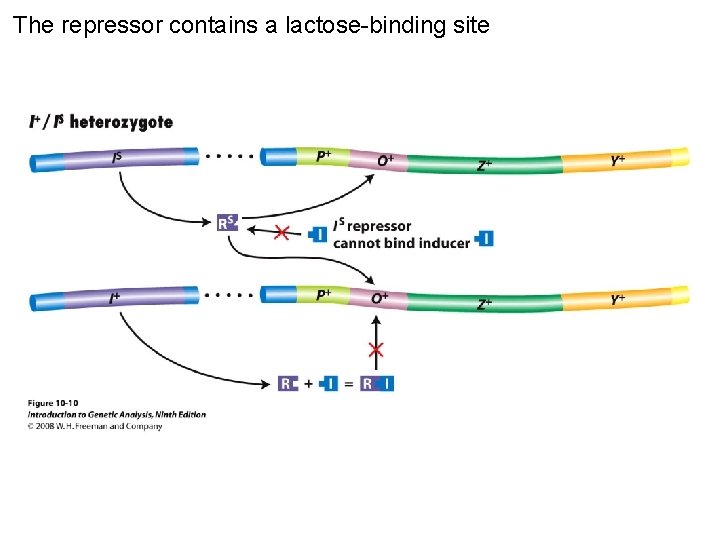
The repressor contains a lactose-binding site Figure 10 -10
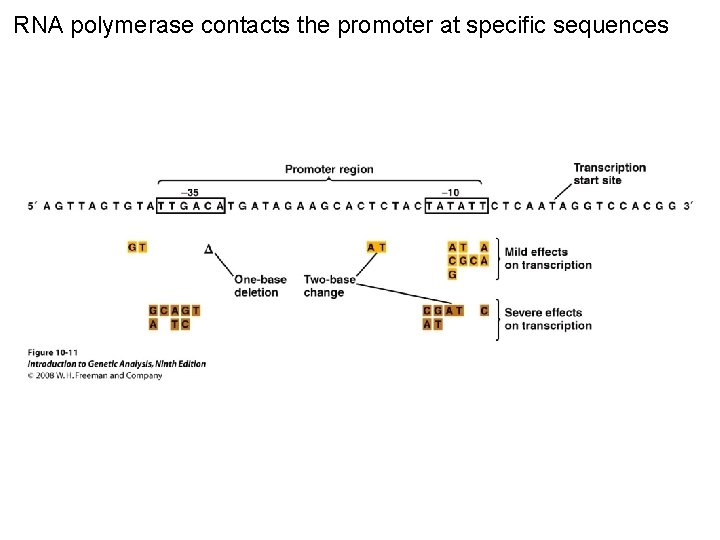
RNA polymerase contacts the promoter at specific sequences Figure 10 -11
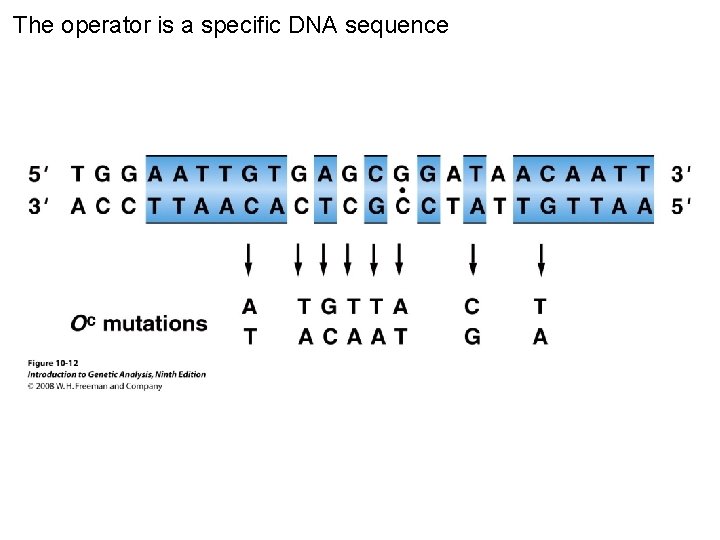
The operator is a specific DNA sequence Figure 10 -12
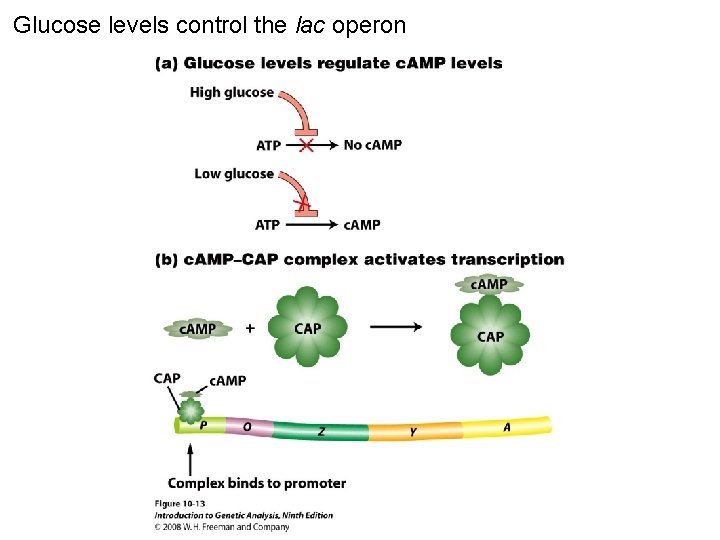
Glucose levels control the lac operon Figure 10 -13
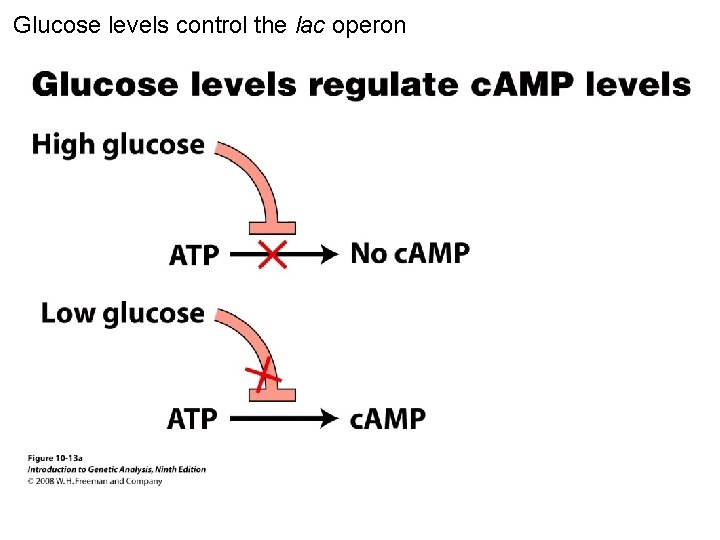
Glucose levels control the lac operon Figure 10 -13 a
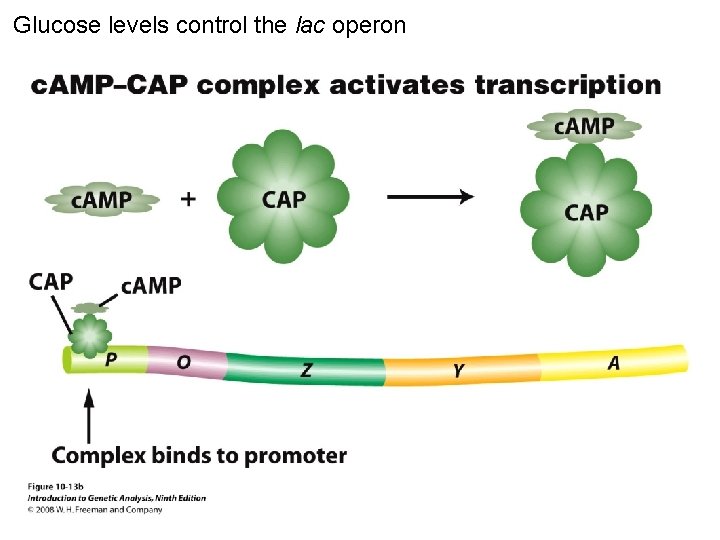
Glucose levels control the lac operon Figure 10 -13 b
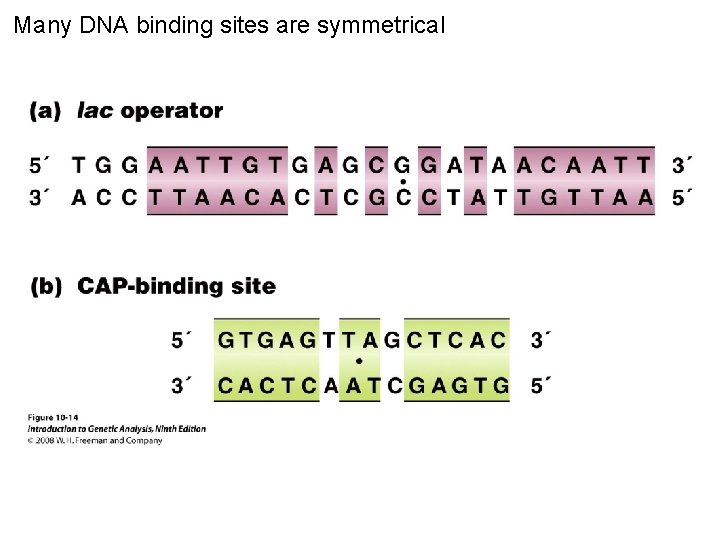
Many DNA binding sites are symmetrical Figure 10 -14
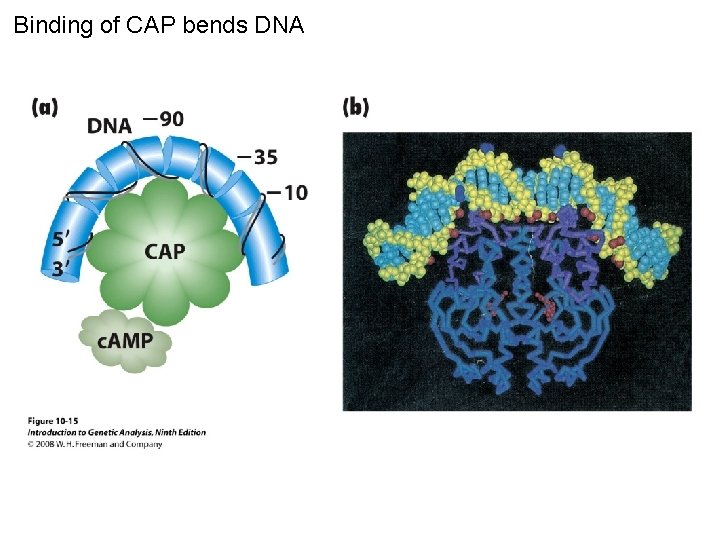
Binding of CAP bends DNA Figure 10 -15
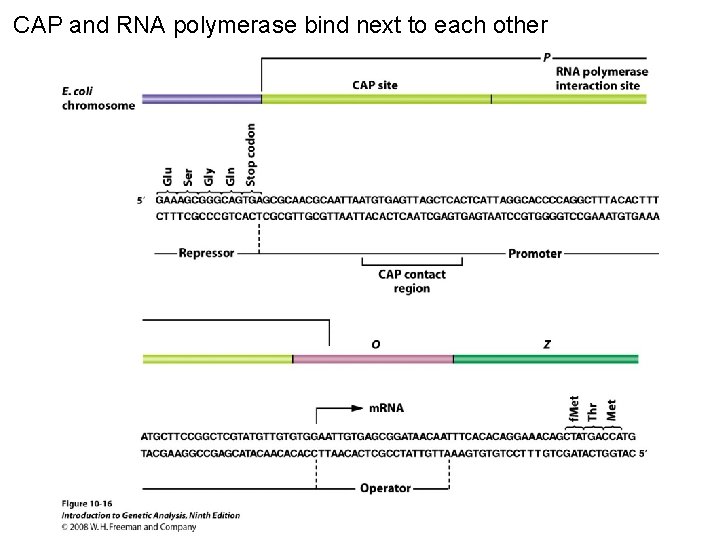
CAP and RNA polymerase bind next to each other Figure 10 -16
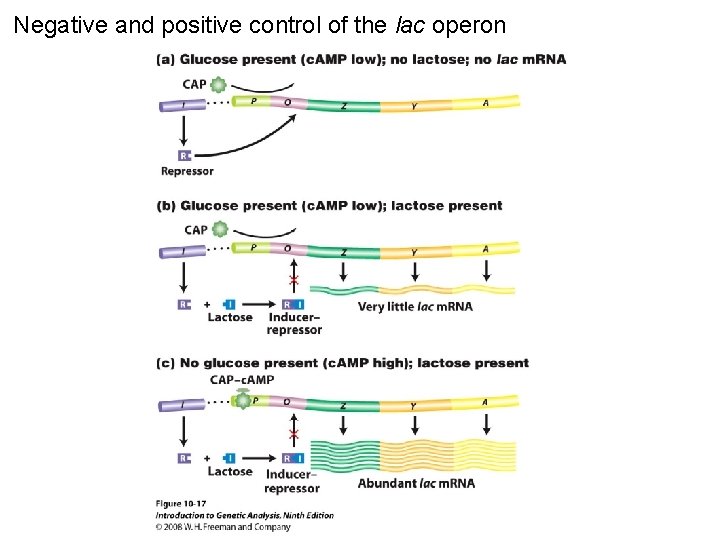
Negative and positive control of the lac operon Figure 10 -17
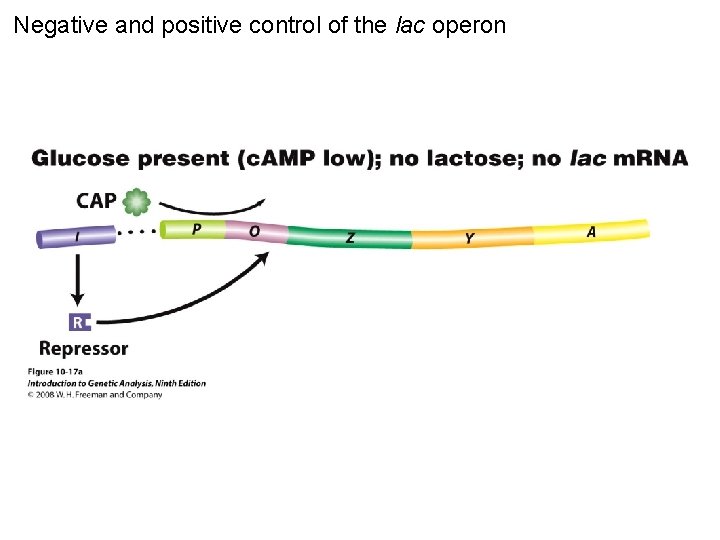
Negative and positive control of the lac operon Figure 10 -17 a
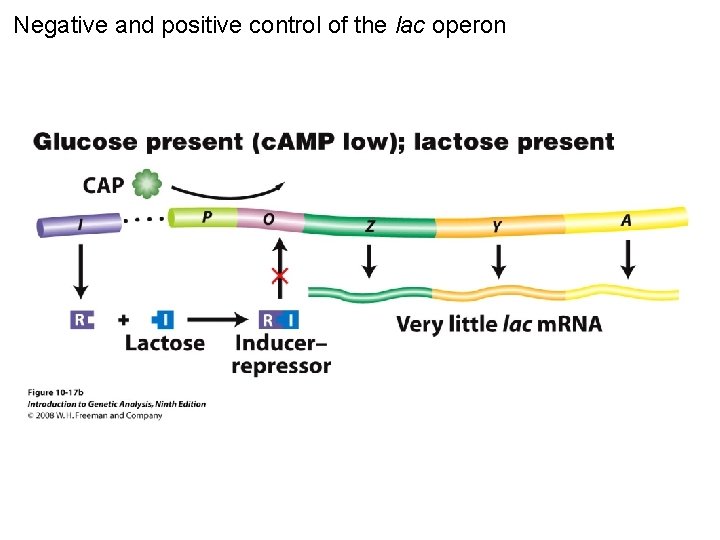
Negative and positive control of the lac operon Figure 10 -17 b
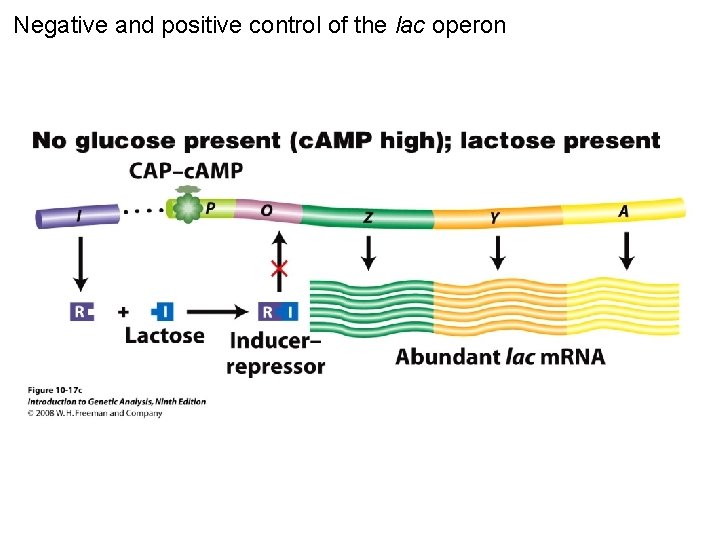
Negative and positive control of the lac operon Figure 10 -17 c
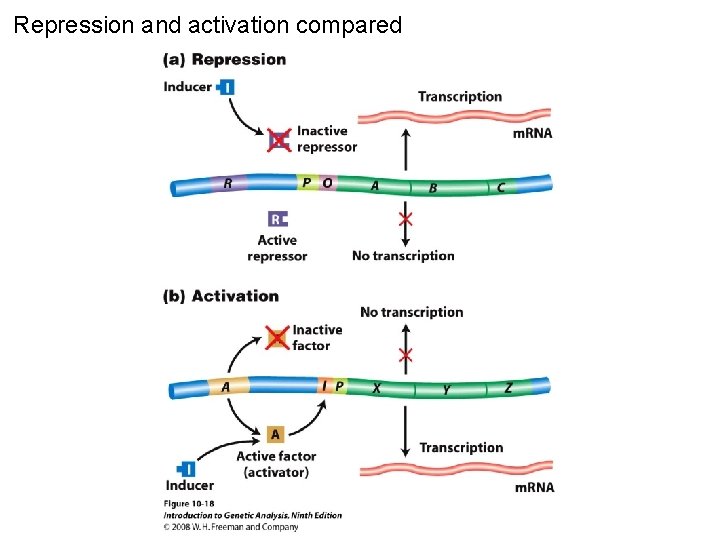
Repression and activation compared Figure 10 -18
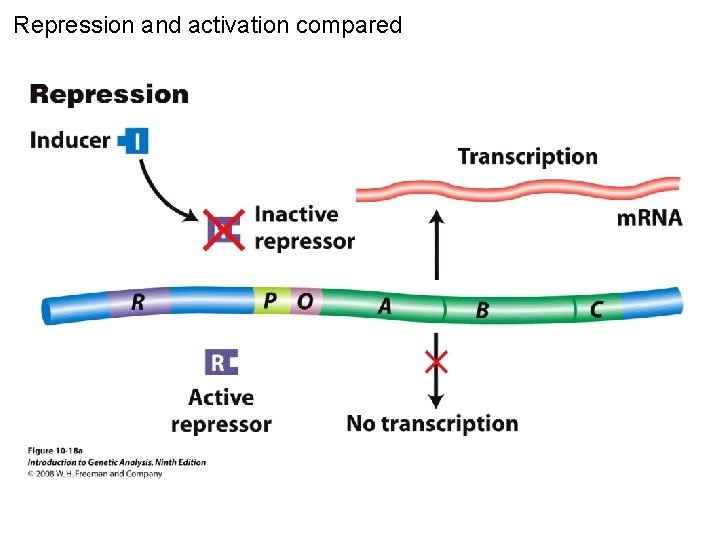
Repression and activation compared Figure 10 -18 a
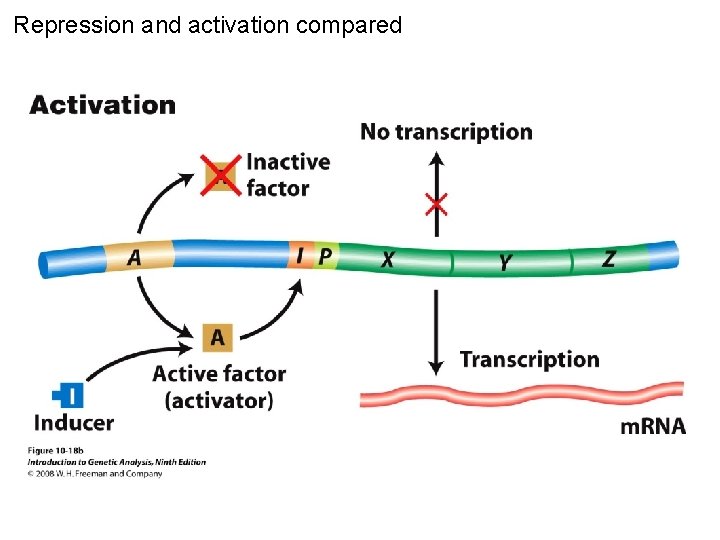
Repression and activation compared Figure 10 -18 b
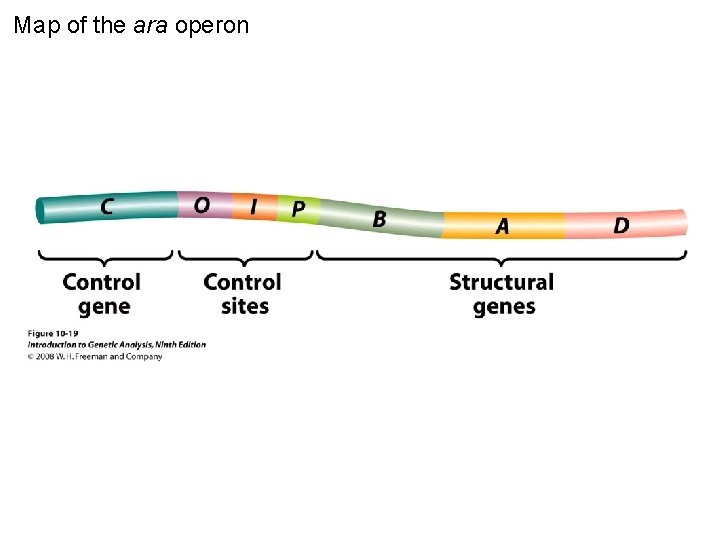
Map of the ara operon Figure 10 -19
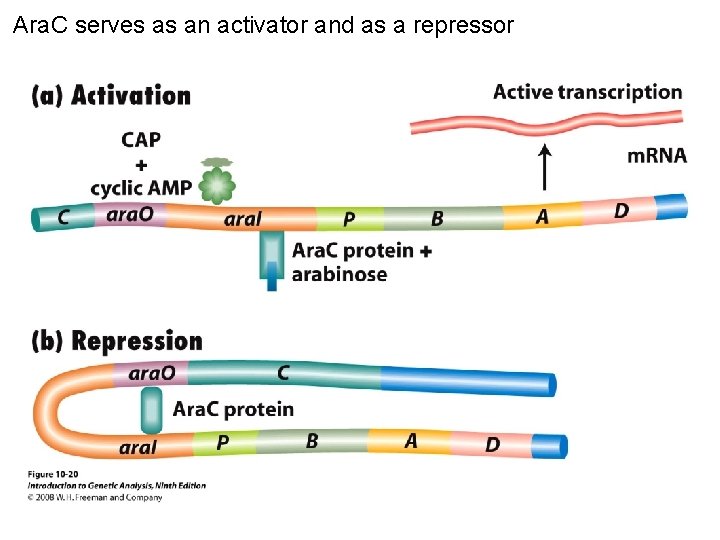
Ara. C serves as an activator and as a repressor Figure 10 -20
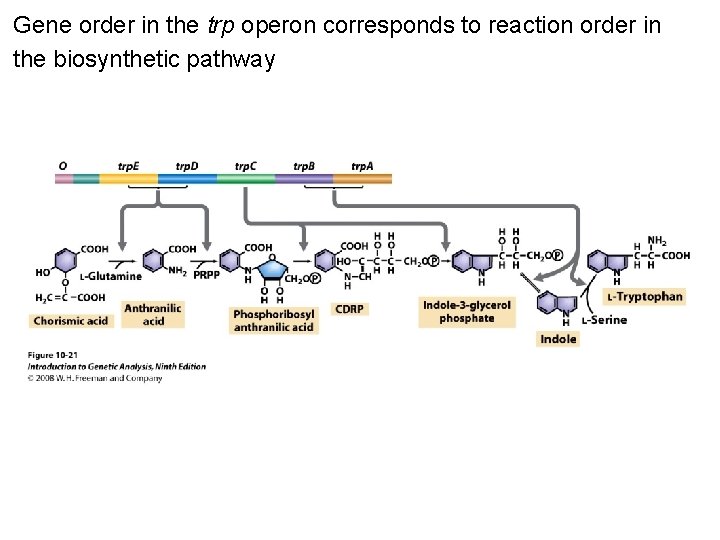
Gene order in the trp operon corresponds to reaction order in the biosynthetic pathway Figure 10 -21
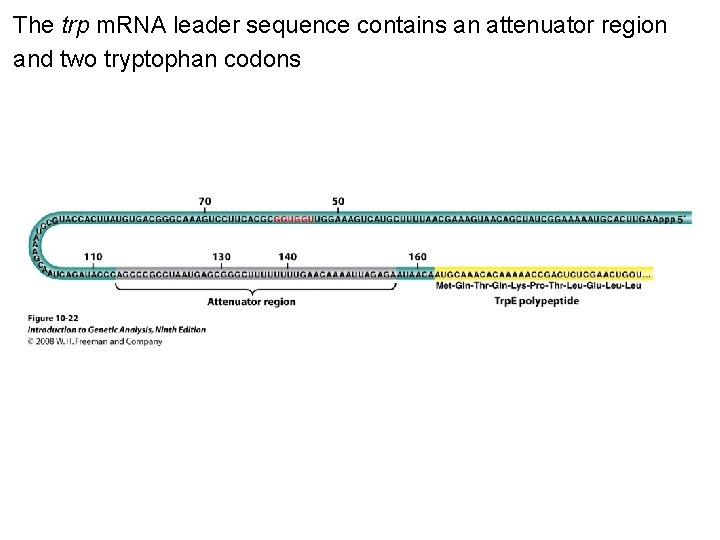
The trp m. RNA leader sequence contains an attenuator region and two tryptophan codons Figure 10 -22
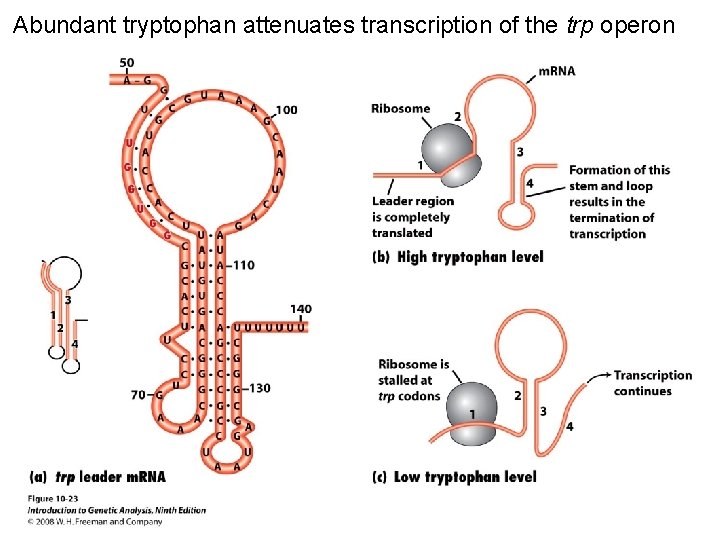
Abundant tryptophan attenuates transcription of the trp operon Figure 10 -23
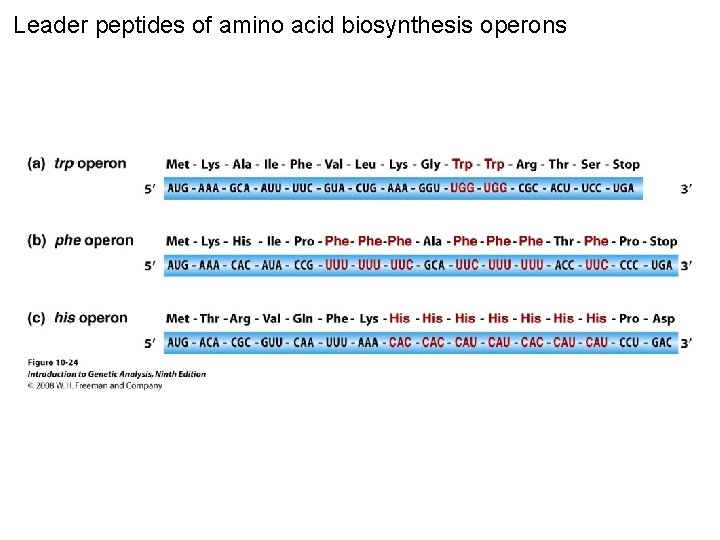
Leader peptides of amino acid biosynthesis operons Figure 10 -24
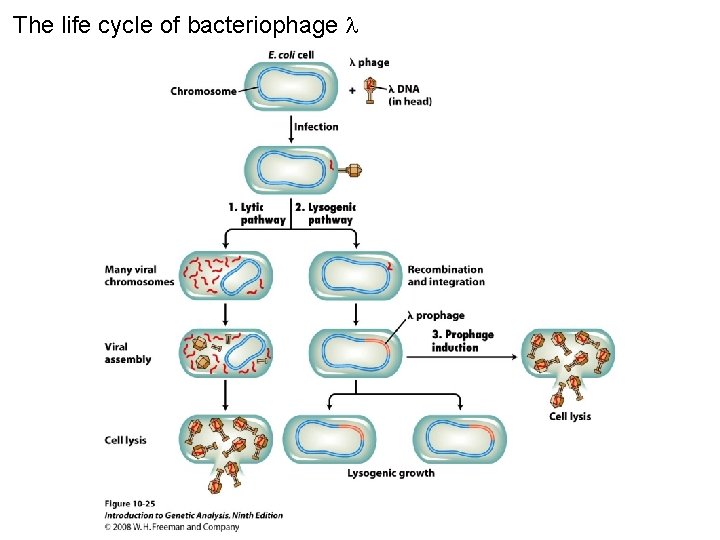
The life cycle of bacteriophage Figure 10 -25
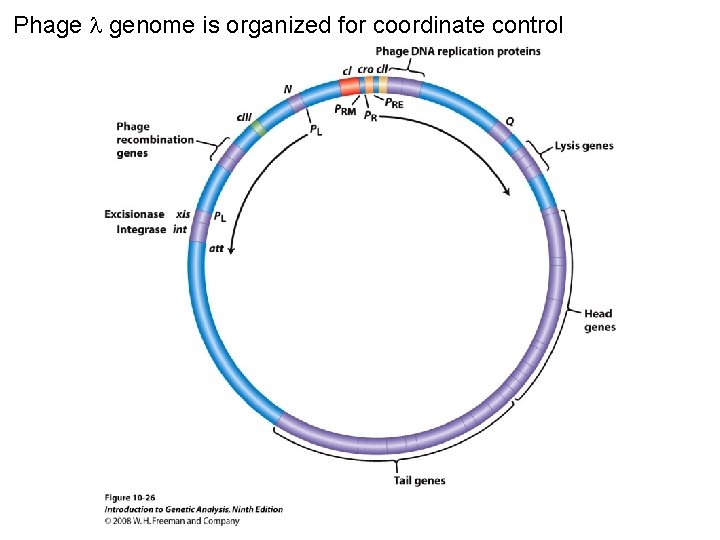
Phage genome is organized for coordinate control Figure 10 -26
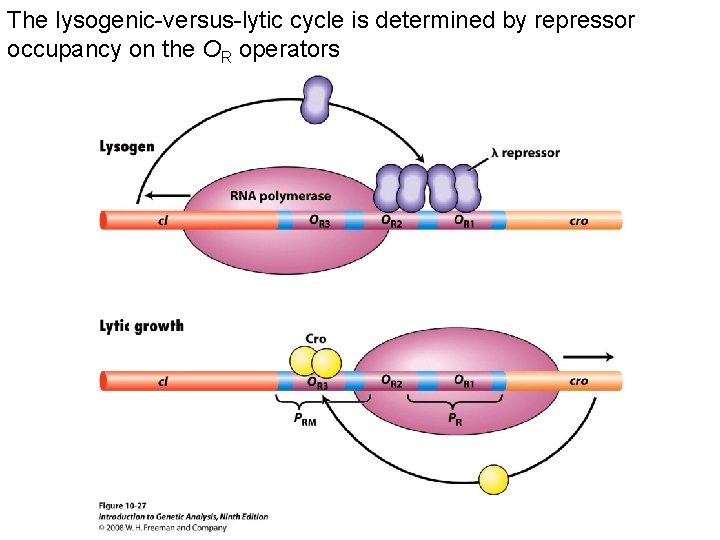
The lysogenic-versus-lytic cycle is determined by repressor occupancy on the OR operators Figure 10 -27
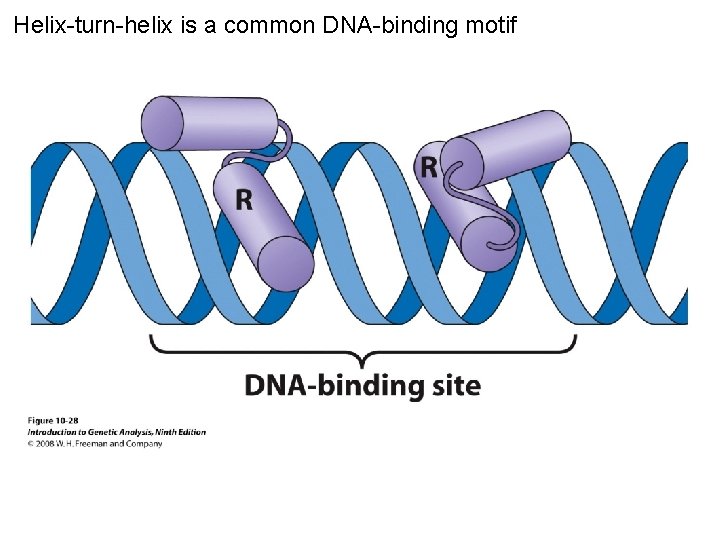
Helix-turn-helix is a common DNA-binding motif Figure 10 -28
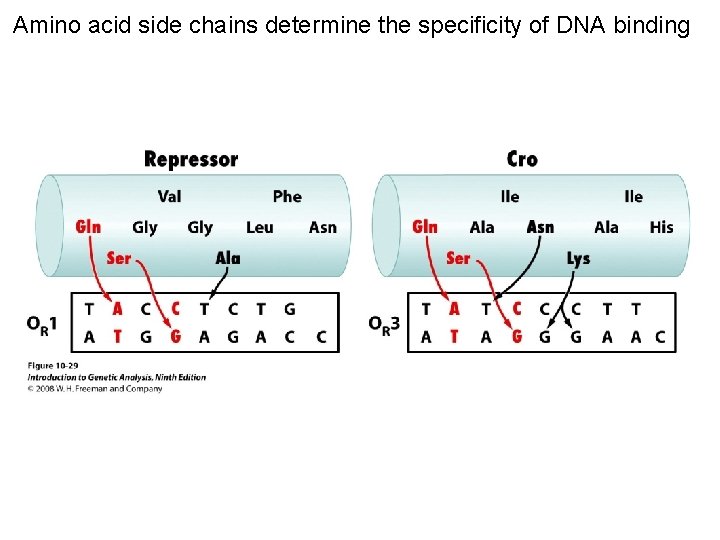
Amino acid side chains determine the specificity of DNA binding Figure 10 -29
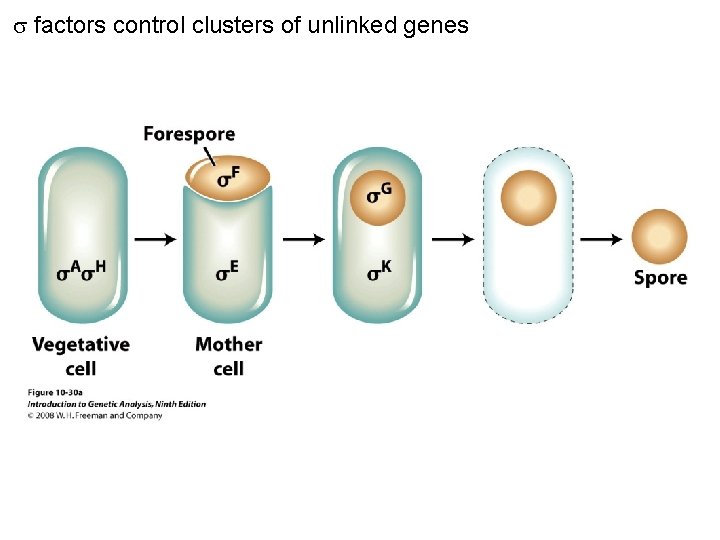
factors control clusters of unlinked genes Figure 10 -30 a
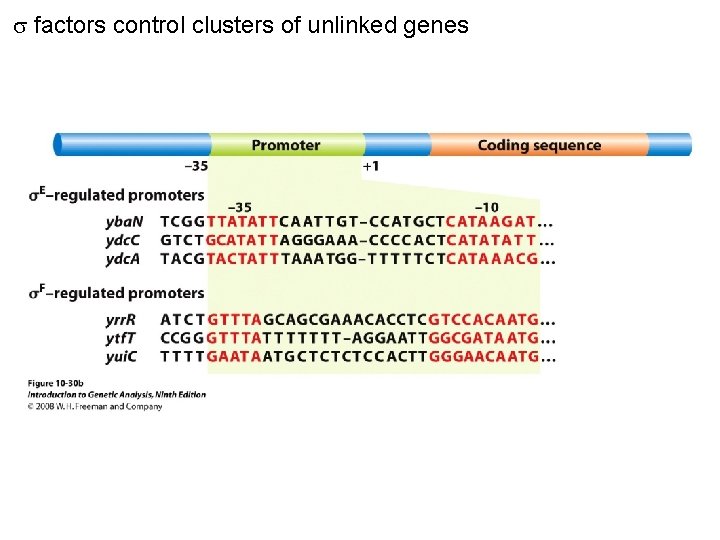
factors control clusters of unlinked genes Figure 10 -30 b
- Slides: 43