Chapter 10 Control of Gene Expression Cengage Learning




- Slides: 39

Chapter 10 Control of Gene Expression © Cengage Learning 2016

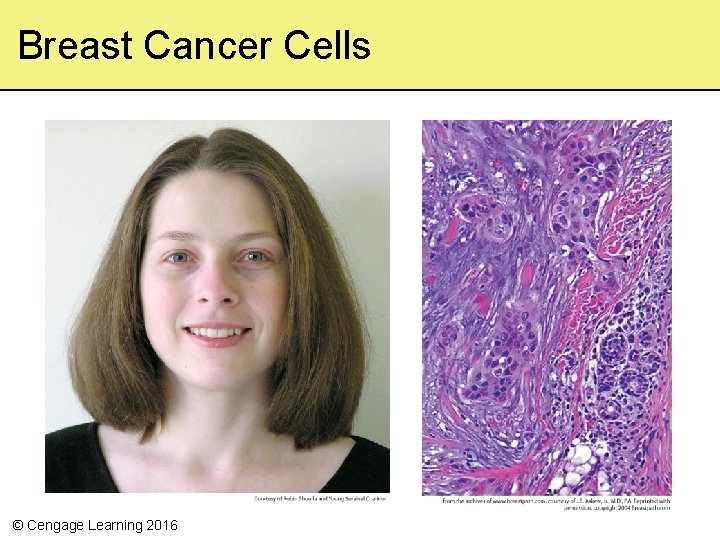
10. 1 Between You and Eternity • Cancer is a multistep process in which cells grow and divide abnormally, disrupting physical and metabolic functions – More than 200, 000 new cases of breast cancer are diagnosed in the US each year – about 5, 700 in women and men under 34 years of age • Mutations in genes that control cell growth and division predispose individuals to develop certain kinds of cancer © Cengage Learning 2016

Breast Cancer Cells © Cengage Learning 2016

10. 2 Switching Genes On and Off • All body cells contain the same DNA with the same genes • Differentiation – The process by which cells become specialized – In multicelled organisms, most cells differentiate when they start expressing a unique subset of their genes – Which genes are expressed depends on the type of organism, its stage of development, and environmental conditions © Cengage Learning 2016

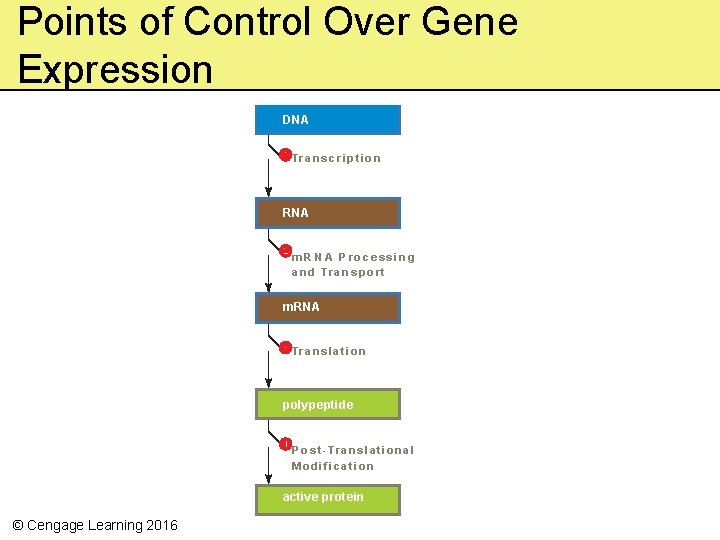
Gene Controls • Govern the kinds and amounts of substances in a cell at any given time • Various control processes regulate all steps between gene and gene product – Gene controls start, enhance, slow, or stop the individual steps of gene expression – Gene controls can operate at any step in the path of protein production © Cengage Learning 2016

Points of Control Over Gene Expression DNA Tr a n s c r i p t i o n RNA m R N A P ro c e s s i n g a n d Tr a n s p o r t m. RNA Tr a n s l a t i o n polypeptide P o s t - Tr a n s l a t i o n a l Modification active protein © Cengage Learning 2016

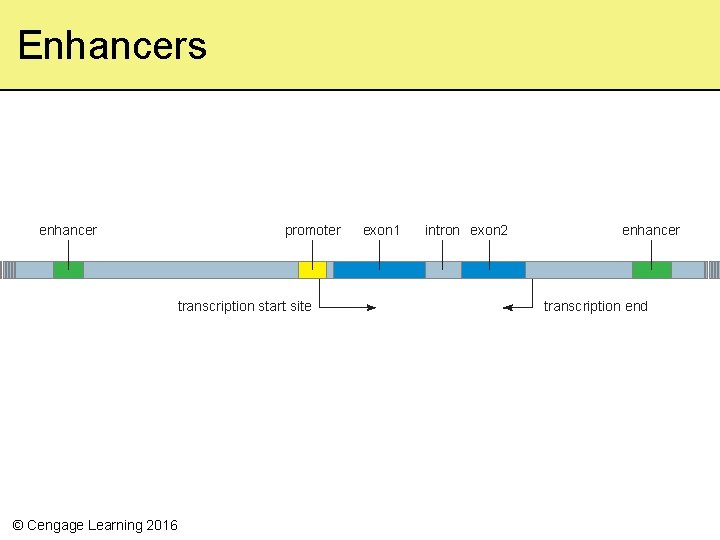
Transcription • Transcription factors – Regulatory proteins that affect the rate of transcription by binding to special nucleotide sequences in DNA – Activators speed up transcription when bound to a promoter; or may bind to distant enhancers – Repressors slow or stop transcription • Chromatin structure also affects transcription © Cengage Learning 2016

Enhancers enhancer promoter transcription start site © Cengage Learning 2016 exon 1 intron exon 2 enhancer transcription end

Control of Transcription • Chemical modifications and chromosome duplications affect RNA polymerase’s access to genes – Enzymes that acetylate histones encourage transcription – Adding a methyl group to a histone prevent transcription – Polytene chromosomes (many copies) increase transcription rates in some organisms © Cengage Learning 2016

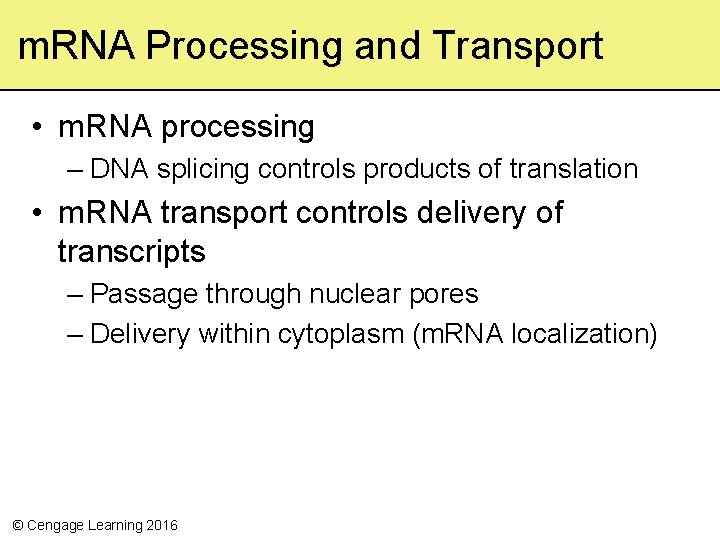
m. RNA Processing and Transport • m. RNA processing – DNA splicing controls products of translation • m. RNA transport controls delivery of transcripts – Passage through nuclear pores – Delivery within cytoplasm (m. RNA localization) © Cengage Learning 2016

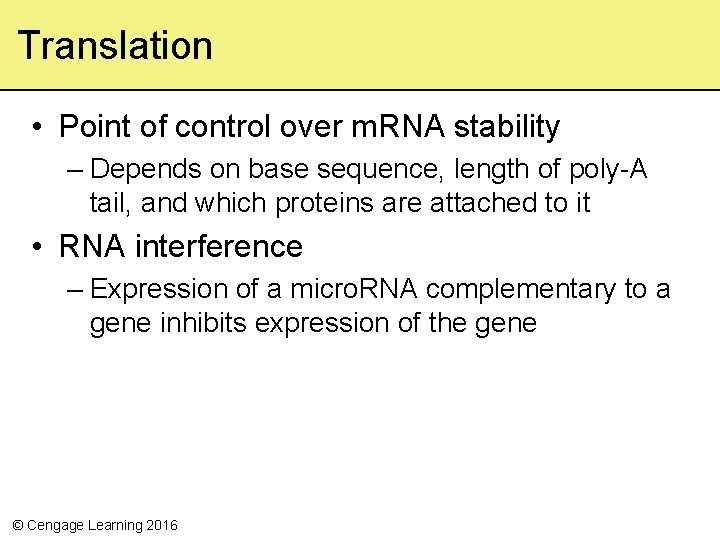
Translation • Point of control over m. RNA stability – Depends on base sequence, length of poly-A tail, and which proteins are attached to it • RNA interference – Expression of a micro. RNA complementary to a gene inhibits expression of the gene © Cengage Learning 2016

Post-Translational Modification • Can inhibit, activate, or stabilize many molecules, including enzymes that participate in transcription and translocation © Cengage Learning 2016

10. 3 Master Genes • Cascades of gene expression govern the development of a complex, multicelled body • Master genes encode products that affect the expression of many other genes • Pattern formation is the process by which a complex body forms from local processes in an embryo © Cengage Learning 2016

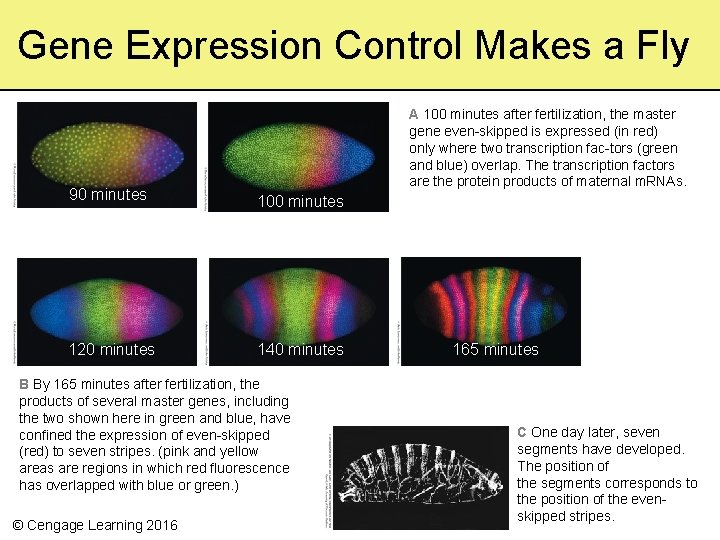
Gene Expression Control Makes a Fly A 100 minutes after fertilization, the master gene even-skipped is expressed (in red) only where two transcription fac-tors (green and blue) overlap. The transcription factors are the protein products of maternal m. RNAs. 90 minutes 100 minutes 120 minutes 140 minutes B By 165 minutes after fertilization, the products of several master genes, including the two shown here in green and blue, have confined the expression of even-skipped (red) to seven stripes. (pink and yellow areas are regions in which red fluorescence has overlapped with blue or green. ) © Cengage Learning 2016 165 minutes C One day later, seven segments have developed. The position of the segments corresponds to the position of the even 13 hours skipped stripes.

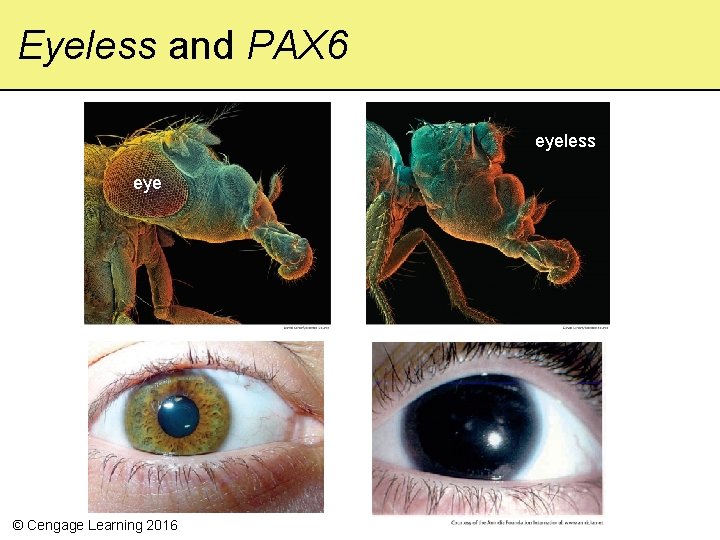
Homeotic Genes • Master genes that control differentiation of specific tissues and body parts in an embryo – Encode transcription factors with a homeodomain • A region of about 60 amino acids that can bind to a promoter – Control development by same mechanisms as in all multicelled eukaryotes • Many are interchangeable among different species © Cengage Learning 2016

Some Effects of Homeotic Gene Mutations © Cengage Learning 2016

Knockout Experiments • Researchers inactivate a gene by introducing a mutation into it, then compare the differences with normal individuals – and similar genes in humans © Cengage Learning 2016

Eyeless and PAX 6 eyeless eye © Cengage Learning 2016 eyeless

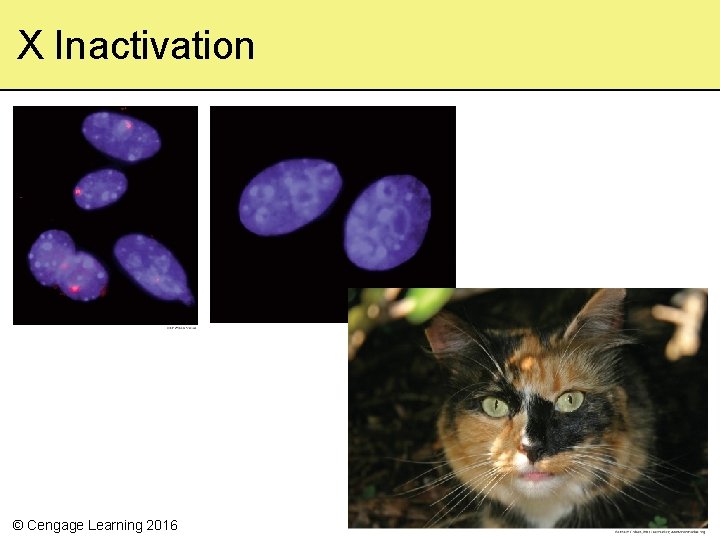
10. 4 Examples of Gene Control in Eukaryotes • Selective gene expression gives rise to many traits • X chromosome inactivation – In cells of female mammals, either the maternal or paternal X chromosome is randomly condensed (Barr body) and is inactive – Occurs in an early embryonic stage, so that all descendants of that particular cell have the same inactive X chromosome, resulting in “mosaic” gene expression © Cengage Learning 2016

Dosage Compensation • The theory that X chromosome inactivation equalizes expression of X chromosome genes between the sexes • Mechanism of X inactivation – XIST gene on one X chromosome transcribes an RNA molecule, which coats the chromosome and causes it to condense, forming a Barr body © Cengage Learning 2016

X Inactivation © Cengage Learning 2016

Male Sex Determination in Humans • Most of the genes on the X chromosome determine nonsexual traits • The Y chromosome carries 307 genes, including SRY – the master gene that triggers formation of testes – Testes produce testosterone that causes formation of male genitalia and secondary sexual traits – In the absence of testosterone, female genitalia form © Cengage Learning 2016

Flower Formation • The ABC model – Three sets of master genes (A, B, C) encode products that initiate cascades of expression of other genes to accomplish intricate tasks such as flower formation – Master genes are expressed differently in tissues of floral shoots – Master genes are switched on by environmental cues such as day length © Cengage Learning 2016

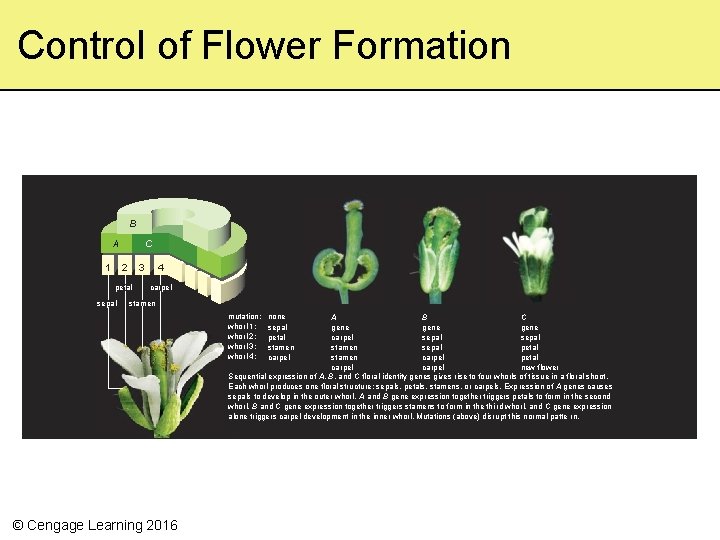
Control of Flower Formation B C A 1 2 3 petal sepal 4 carpel stamen mutation: whorl 1: whorl 2: whorl 3: whorl 4: none sepal petal stamen carpel A B C gene carpel sepal stamen sepal petal stamen carpel petal carpel new flower Sequential expression of A, B, and C floral identity genes gives rise to four whorls of tissue in a floral shoot. Each whorl produces one floral structure: sepals, petals, stamens, or carpels. Exp ression of A genes causes sepals to develop in the outer whorl. A and B gene expression together triggers petals to form in the second whorl. B and C gene expression together triggers stamens to form in the thi rd whorl, and C gene expression alone triggers carpel development in the inner whorl. Mutations (above) disrupt this normal patte rn. © Cengage Learning 2016

10. 5 Gene Control in Prokaryotes • Prokaryotes (bacteria and archaea) are single-celled and do not have master genes • Prokaryotes control gene expression mainly by adjusting the rate of transcription in response to shifts in nutrient availability and other outside conditions © Cengage Learning 2016

Operons and Operators • In prokaryotes, genes that are used together often occur together on chromosomes • Operon – A promoter and one or more operators that collectively control transcription of multiple genes • Operators – DNA regions that are binding sites for a repressor © Cengage Learning 2016

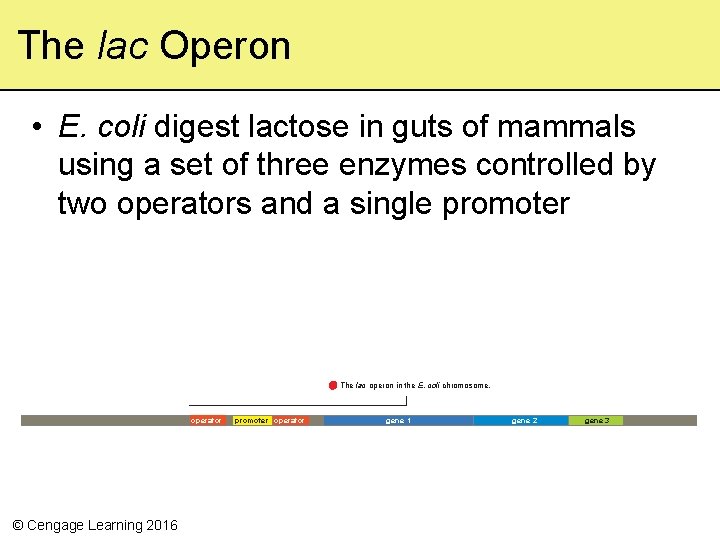
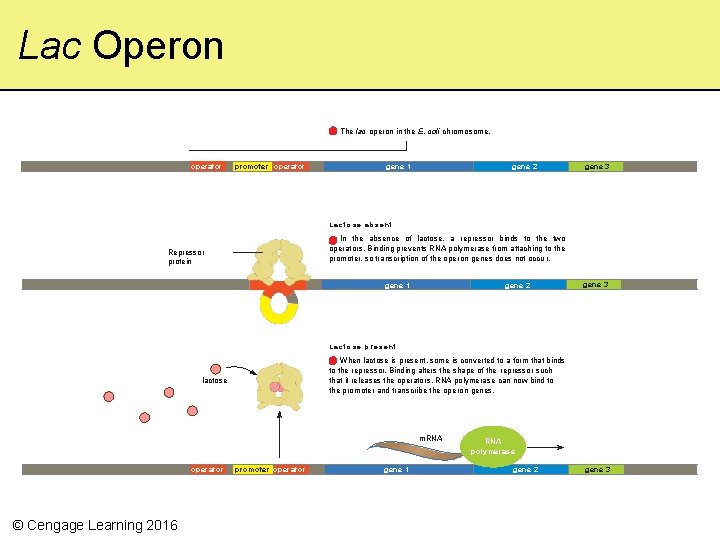
The lac Operon • E. coli digest lactose in guts of mammals using a set of three enzymes controlled by two operators and a single promoter The lac operon in the E. coli chromosome. operator © Cengage Learning 2016 promoter operator gene 1 gene 2 gene 3

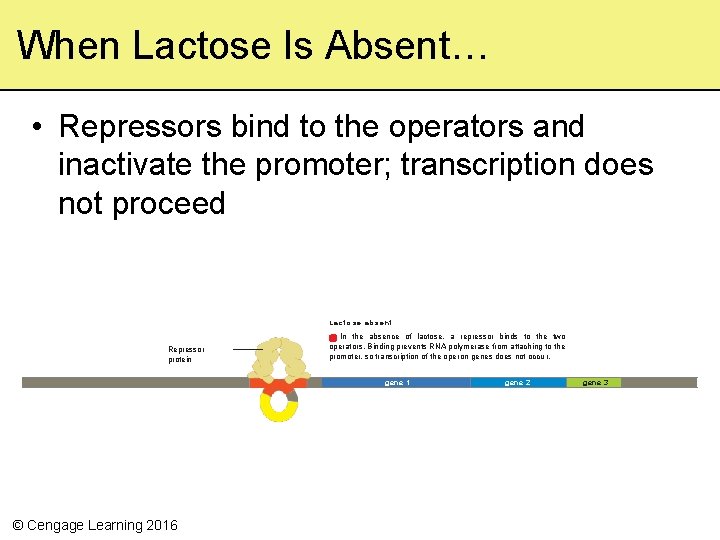
When Lactose Is Absent… • Repressors bind to the operators and inactivate the promoter; transcription does not proceed Lactose absent Repressor protein In the absence of lactose, a repressor binds to the two operators. Binding prevents RNA polymerase from attaching to the promoter, so transcription of the ope ron genes does not occu r. gene 1 © Cengage Learning 2016 gene 2 gene 3

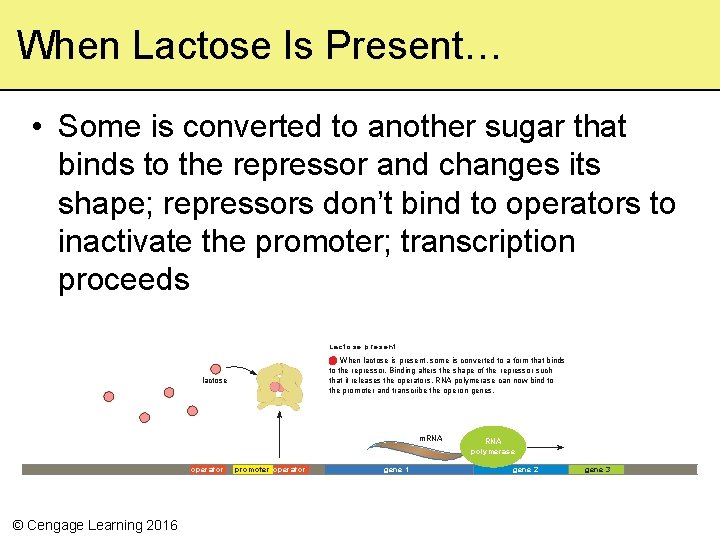
When Lactose Is Present… • Some is converted to another sugar that binds to the repressor and changes its shape; repressors don’t bind to operators to inactivate the promoter; transcription proceeds L a c t o s e p re s e n t When lactose is present, some is converted to a form that binds to the repressor. Binding alters the shape of the repressor such that it releases the operators. RNA polymerase can now bind to the promoter and transcribe the ope ron genes. lactose m. RNA polymerase operator © Cengage Learning 2016 promoter operator gene 1 gene 2 gene 3

Lac Operon The lac operon in the E. coli chromosome. operator promoter operator gene 1 gene 2 gene 3 Lactose absent In the absence of lactose, a repressor binds to the two operators. Binding prevents RNA polymerase from attaching to the promoter, so transcription of the ope ron genes does not occu r. Repressor protein gene 1 gene 2 gene 3 L a c t o s e p re s e n t When lactose is present, some is converted to a form that binds to the repressor. Binding alters the shape of the repressor such that it releases the operators. RNA polymerase can now bind to the promoter and transcribe the ope ron genes. lactose m. RNA polymerase operator © Cengage Learning 2016 promoter operator gene 1 gene 2 gene 3

Lactose Intolerance • Human infants and other mammals produce the enzyme lactase, which digests the lactose in milk • The majority of humans begin to lose the ability to produce lactase around age 5, and become lactose intolerant • Many people of European ancestry carry a mutation of one of the genes responsible for programmed lactase shutdown © Cengage Learning 2016

Riboswitches • Some bacterial m. RNAs regulate their own translation with riboswitches – small sequences of RNA nucleotides that bind to a target molecule • Binding of an end product (such as vitamin B 12) changes the shape of the m. RNA so that ribosomes no longer attach to it, and translation stops – an example of feedback inhibition © Cengage Learning 2016

10. 6 Epigenetics • Methylations and other modifications that accumulate in DNA during an individual’s lifetime can be passed to offspring – Methylation in the parental DNA is usually reset but not always – This is epigenetic inheritance that does not modify the nucleotide sequence © Cengage Learning 2016

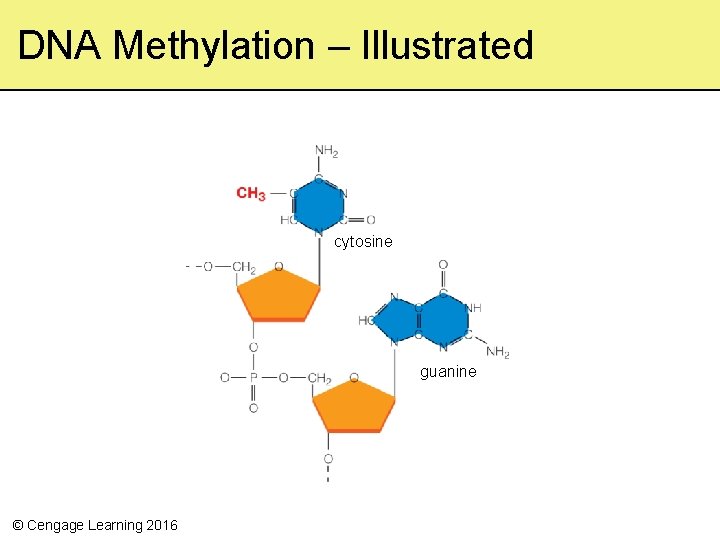
DNA Methylations • Between 3 and 6 percent of DNA in body cells is methylated • Methyl groups often attach to a cytosine followed by a guanine, but which cytosines are methylated varies by individual • In some cases, a decrease in methylations that results in an increase in expression of a gene may offer a survival advantage © Cengage Learning 2016

DNA Methylation – Illustrated cytosine guanine © Cengage Learning 2016

Heritable Methylations • Once a base in a cell’s DNA becomes methylated, it usually stays methylated in all of the cell’s descendants – Methylation patterns in parental DNA is normally “reset”, with new methyl groups being added and old ones being removed – However, not all parental methyl groups are removed, so methylations acquired during an individual’s lifetime can be passed to offspring © Cengage Learning 2016

Epigenetic Inheritance • Any heritable changes in gene expression that are not due to changes in DNA sequence are said to be epigenetic – Epigenetic inheritance can adapt offspring to environmental stressors much more quickly than evolutionary processes – Epigenetic marks may persist for generations after an environmental stressor has faded © Cengage Learning 2016

Epigenetic Inheritance Is Sex-Limited • Grandsons of boys who endured a winter of famine when they were 6 years old lived about 32 years longer than the grandsons of boys who overate at the same age • Effects are sex-limited – Boys are affected by lifestyle of male ancestors; – Girls by individuals in the maternal line © Cengage Learning 2016

Points to Ponder • What are the advantages and disadvantages of learning you have a gene that may lead to cancer? • Why is gene control research more difficult among eukaryotic species than it is among prokaryotic species? • How might alterations in DNA structure be harmful or beneficial to a species? © Cengage Learning 2016