Chapter 10 Classification 2004 Wadsworth Thomson Learning Major



- Slides: 14

Chapter 10 Classification © 2004 Wadsworth – Thomson Learning

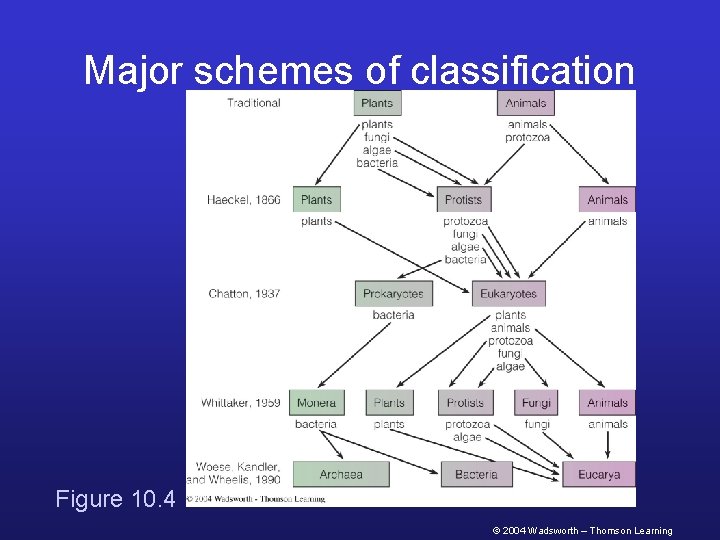
Major schemes of classification Figure 10. 4 © 2004 Wadsworth – Thomson Learning

Biological classification • • Domain Kingdom Phylum Class Order Family Genus Species © 2004 Wadsworth – Thomson Learning

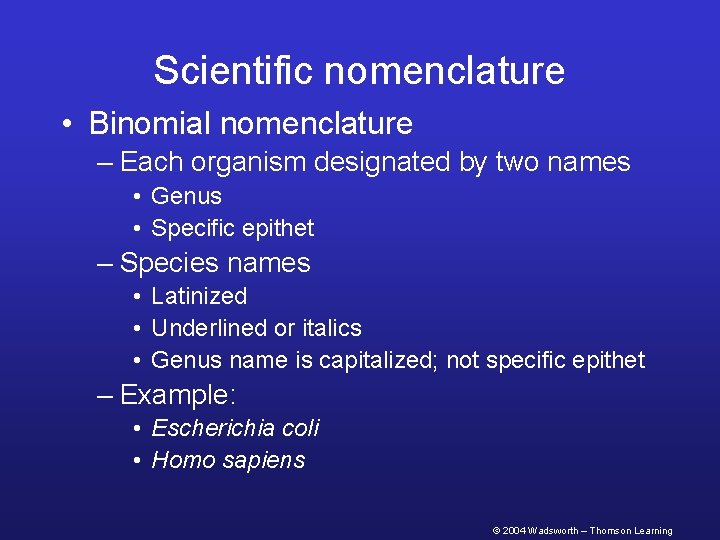
Scientific nomenclature • Binomial nomenclature – Each organism designated by two names • Genus • Specific epithet – Species names • Latinized • Underlined or italics • Genus name is capitalized; not specific epithet – Example: • Escherichia coli • Homo sapiens © 2004 Wadsworth – Thomson Learning

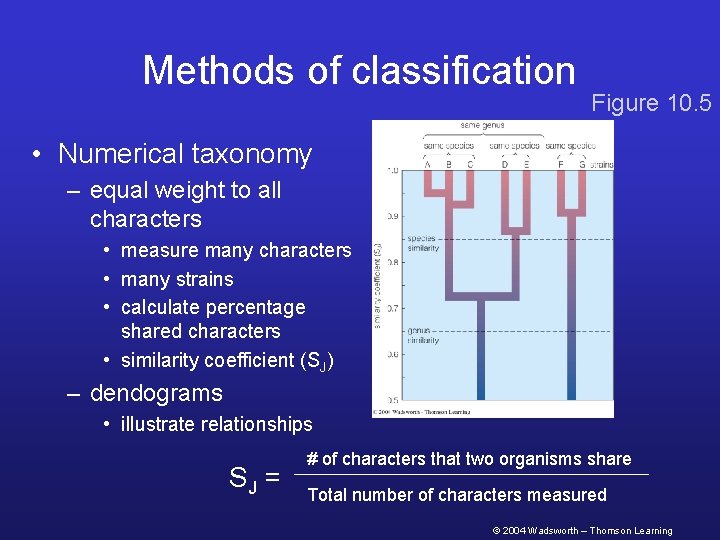
Methods of classification Figure 10. 5 • Numerical taxonomy – equal weight to all characters • measure many characters • many strains • calculate percentage shared characters • similarity coefficient (SJ) – dendograms • illustrate relationships SJ = # of characters that two organisms share Total number of characters measured © 2004 Wadsworth – Thomson Learning

Traditional Methods • Morphology – shape • cocci, bacilli, spirilla – arrangement • singly, chains, clusters – flagella arrangement – Gram reaction • gram-positive, gram-negative © 2004 Wadsworth – Thomson Learning

Traditional Methods • Biochemical and Physiological – growth requirements – end products – enzymes produced – carbon source • oxidation/reduction measurement • tetrazolium dye – fermentation properties • acids, gases © 2004 Wadsworth – Thomson Learning

Traditional Methods • Serology – antibodies • highly specific for target molecules • distinguish between closely related – strain differentiation • Phage typing – viruses that infect bacteria • pattern of strains attacked by specific phage • host range of phage is narrow © 2004 Wadsworth – Thomson Learning

Genomic methods: Percent G+C • Fraction of guanine-cytosine pairs – melting point of DNA – density measurements • % G+C indicate relatedness – values vary – similarities do not prove relatedness © 2004 Wadsworth – Thomson Learning

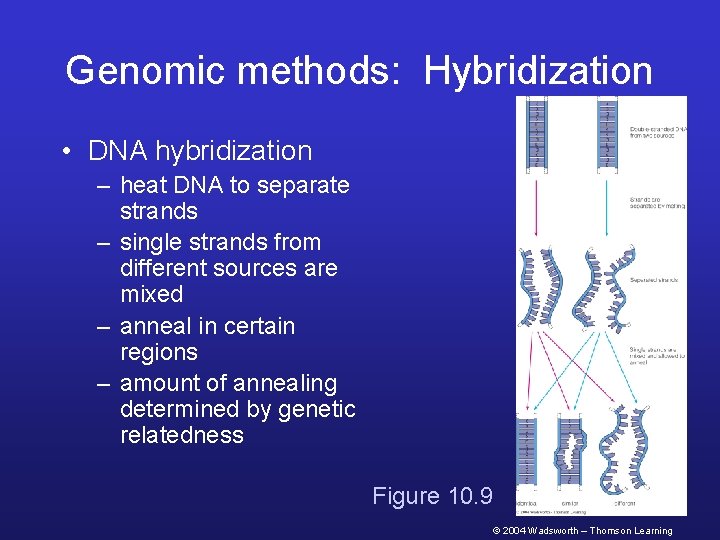
Genomic methods: Hybridization • DNA hybridization – heat DNA to separate strands – single strands from different sources are mixed – anneal in certain regions – amount of annealing determined by genetic relatedness Figure 10. 9 © 2004 Wadsworth – Thomson Learning

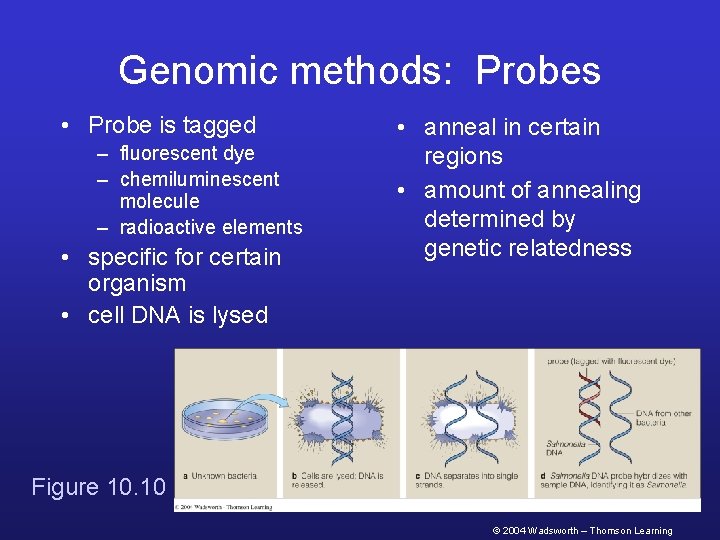
Genomic methods: Probes • Probe is tagged – fluorescent dye – chemiluminescent molecule – radioactive elements • specific for certain organism • cell DNA is lysed • anneal in certain regions • amount of annealing determined by genetic relatedness Figure 10. 10 © 2004 Wadsworth – Thomson Learning

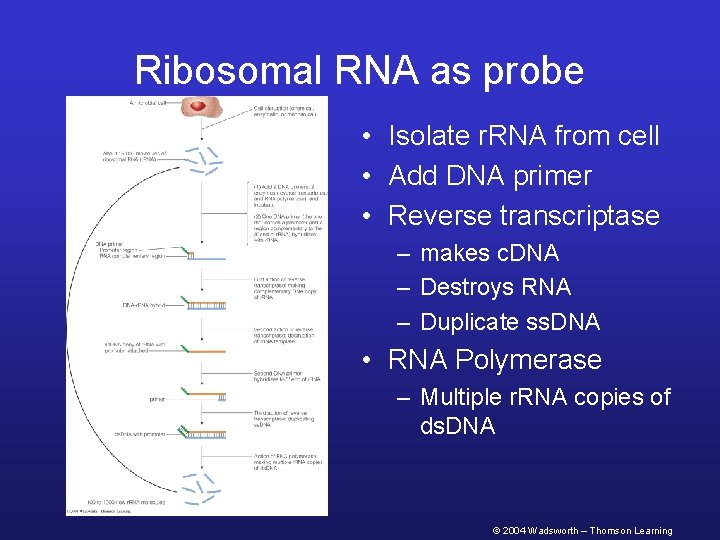
Ribosomal RNA as probe • Isolate r. RNA from cell • Add DNA primer • Reverse transcriptase – makes c. DNA – Destroys RNA – Duplicate ss. DNA • RNA Polymerase – Multiple r. RNA copies of ds. DNA © 2004 Wadsworth – Thomson Learning

Genomic methods: Sequence • DNA Sequencing – ultimate tool of taxonomy • genome sequencing most accurate • formidable task--large size of genomes – 4. 5 million base pairs in E. coli – Ribosomal RNA genes • • small ribosomal subunit all cellular organisms contain r. RNA evolves more slowly highly conserved – Protein encoding genes © 2004 Wadsworth – Thomson Learning

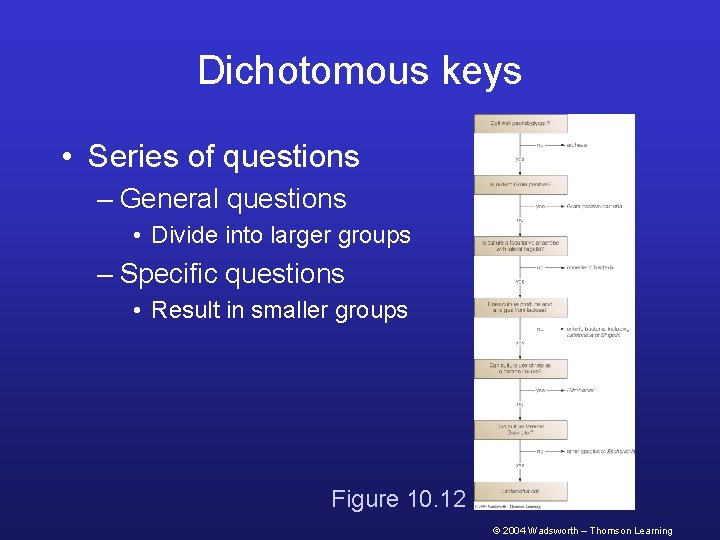
Dichotomous keys • Series of questions – General questions • Divide into larger groups – Specific questions • Result in smaller groups Figure 10. 12 © 2004 Wadsworth – Thomson Learning