CHAPTER 10 Biotechnology Techniques Chapter Outline 10 1

CHAPTER 10: Biotechnology Techniques

Chapter Outline 10 -1 Purification and Detection of Nucleic Acids 10 -2 Restriction Endonucleases 10 -3 Cloning 10 -4 Genetic Engineering 10 -5 DNA Libraries 10 -6 The Polymerase Chain Reaction 10 -7 DNA Fingerprinting 10 -8 Sequencing DNA 10 -9 Genomics and Proteomics
Purification and Detection of Nucleic Acids • Gel electrophoresis is a common technique used to separate nucleic acids. • Based on motion of charged particles in an electric field
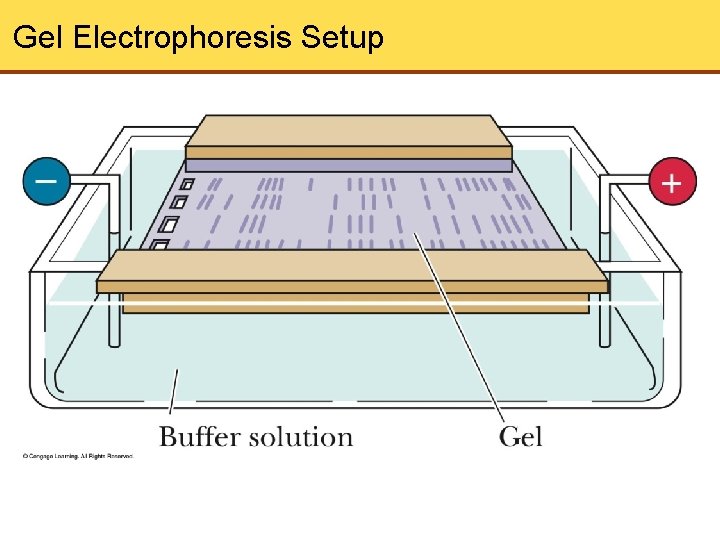
Gel Electrophoresis Setup
Purification and Detection (Cont’d) • Radioactive labeling of sample used to detect products • Label or tag allows visualization • DNA undergo reaction that incorporate radioactive isotope into the DNA • Autoradiography used to visualize image that has been exposed to oligonucleotides that have been radiolabeled • Fluorescence also used. Ethidium Bromide…can slip between DNA bases, and it has different fluorescence characteristics as opposed to when it is free in solution • Et. Br is used as stain for DNA on gels. Et. Br is dangerous ( a carcinogen)…new fluorescent dyes have been developed (Sy. Br Green and Gold)
Restriction Endonucleases • Nucleases- catalyze the hydrolysis of the phosphodiester backbone of nucleic acids - Endonuclease: cleavage in the middle of the chain - Exonuclease: cleavage from the ends of the molecule • Restriction Endonucleases- Have a crucial role in development of recombinant DNA technology • Bacteriophages, viruses that infect bacteria, were being studied when restriction enzymes were discovered
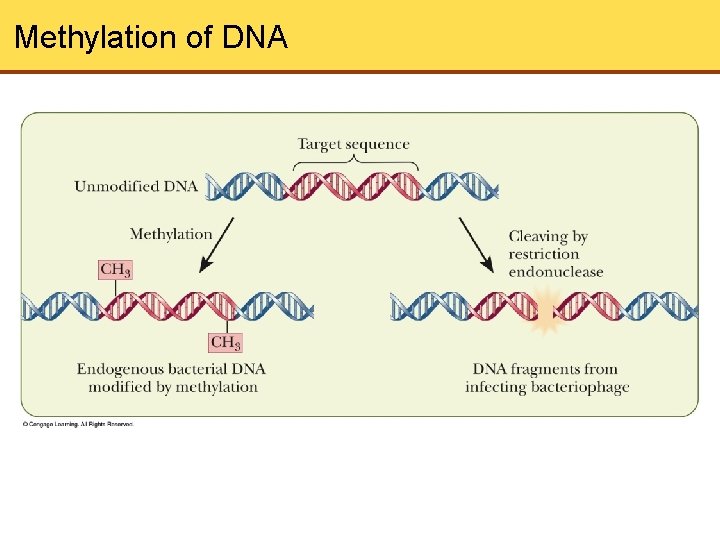
Methylation of DNA
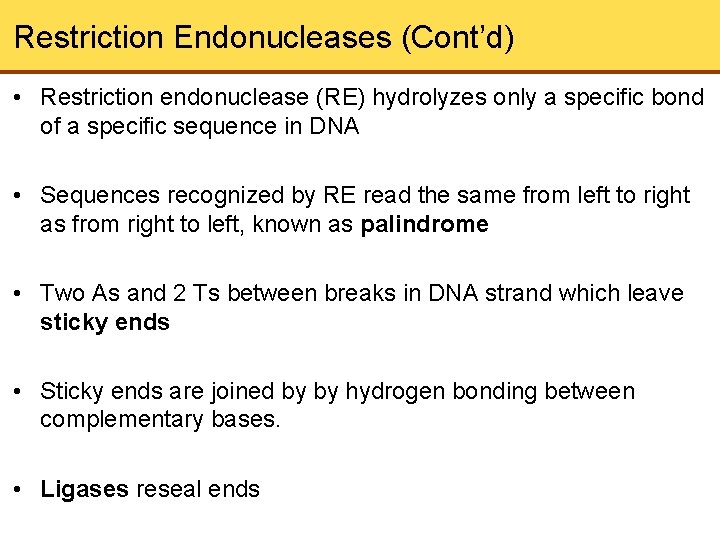
Restriction Endonucleases (Cont’d) • Restriction endonuclease (RE) hydrolyzes only a specific bond of a specific sequence in DNA • Sequences recognized by RE read the same from left to right as from right to left, known as palindrome • Two As and 2 Ts between breaks in DNA strand which leave sticky ends • Sticky ends are joined by by hydrogen bonding between complementary bases. • Ligases reseal ends
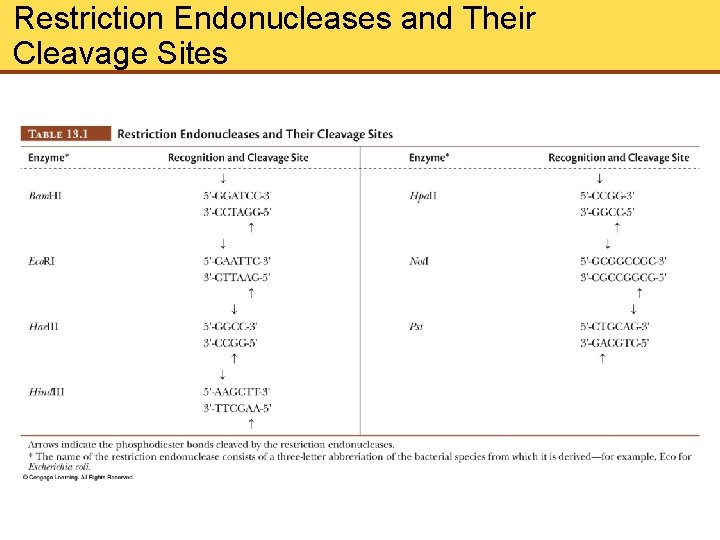
Restriction Endonucleases and Their Cleavage Sites
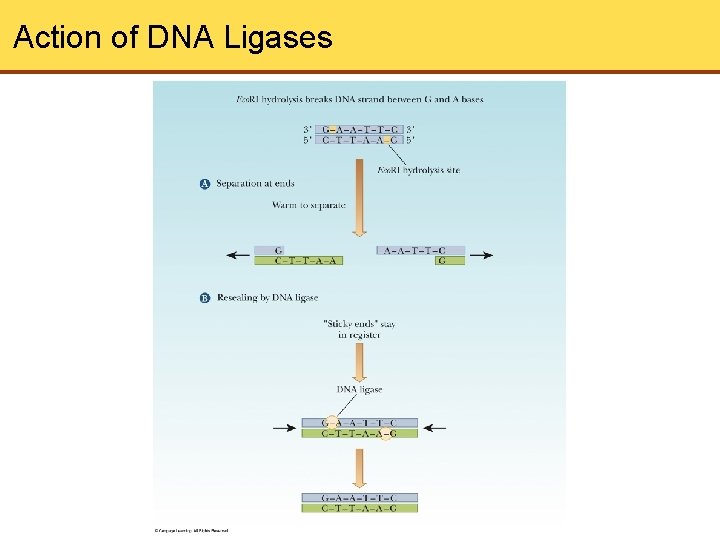
Action of DNA Ligases
Cloning • Recombinant DNA- DNA molecules that contain covalently linked segments derived from 2 or more DNA sources • Sticky Ends can be used to construct Recombinant DNA • DNA Ligase- seals nicks in the covalent structure • Plasmid- small circular DNA that is not part of the main circular DNA chromosome of the bacterium. • Cloning- The process of making identical copies of DNA
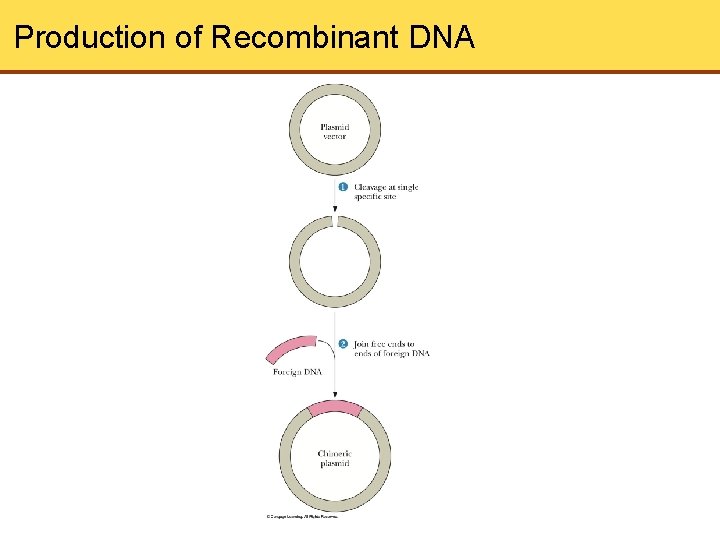
Production of Recombinant DNA
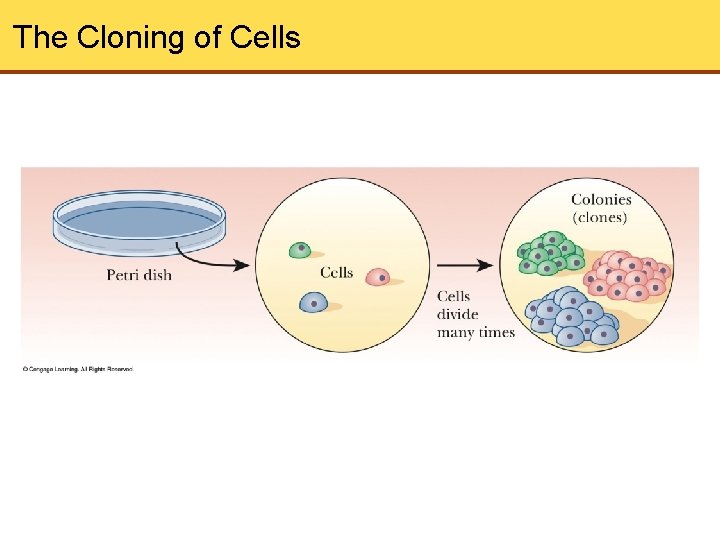
The Cloning of Cells
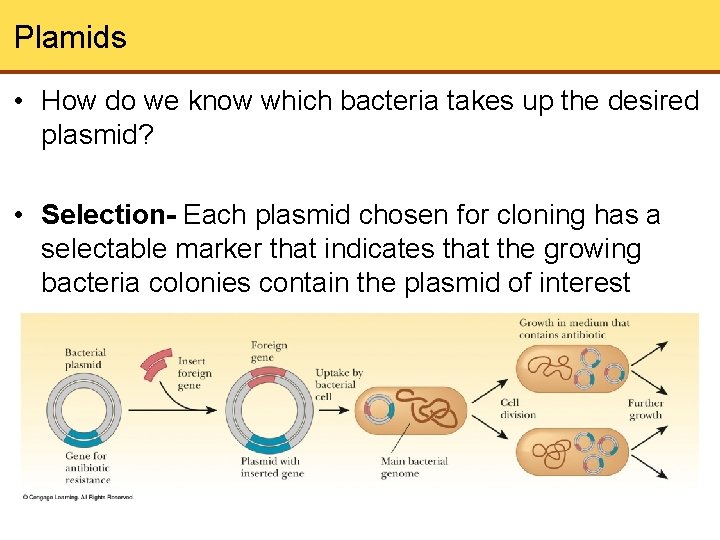
Plamids • How do we know which bacteria takes up the desired plasmid? • Selection- Each plasmid chosen for cloning has a selectable marker that indicates that the growing bacteria colonies contain the plasmid of interest
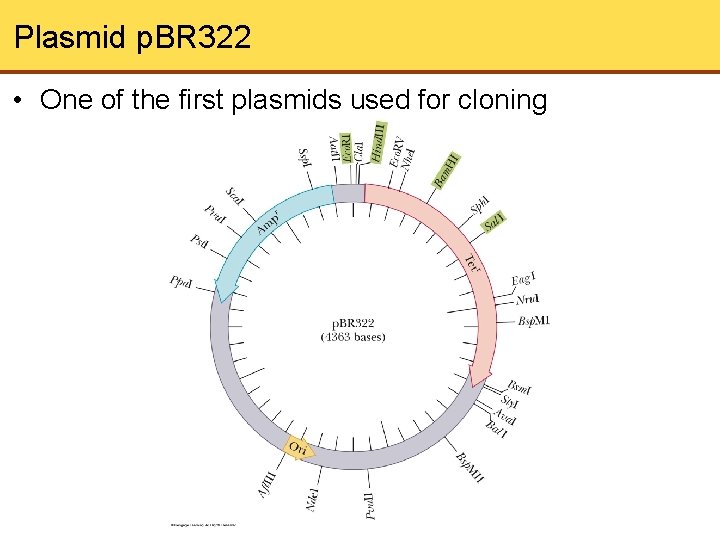
Plasmid p. BR 322 • One of the first plasmids used for cloning
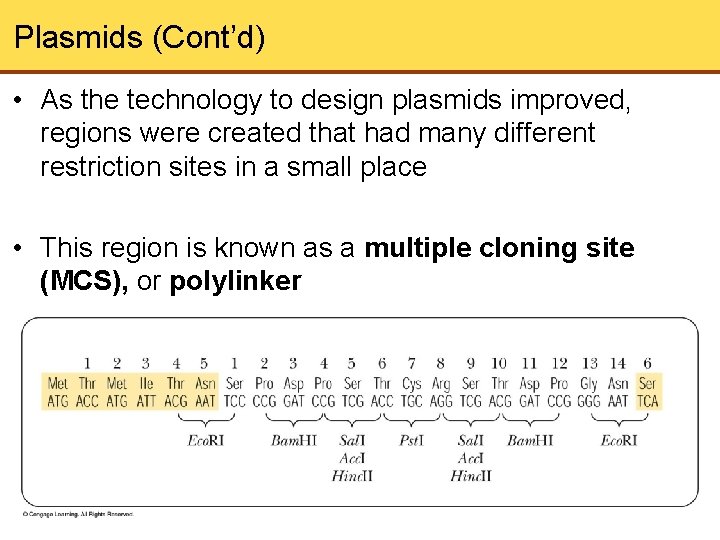
Plasmids (Cont’d) • As the technology to design plasmids improved, regions were created that had many different restriction sites in a small place • This region is known as a multiple cloning site (MCS), or polylinker
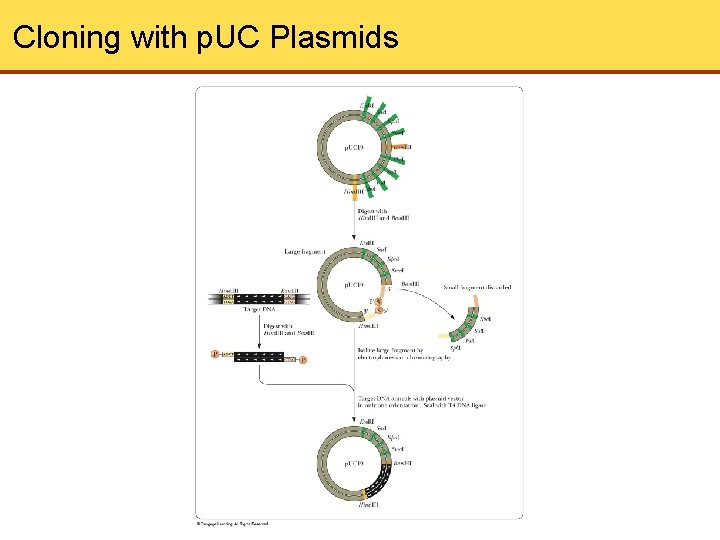
Cloning with p. UC Plasmids
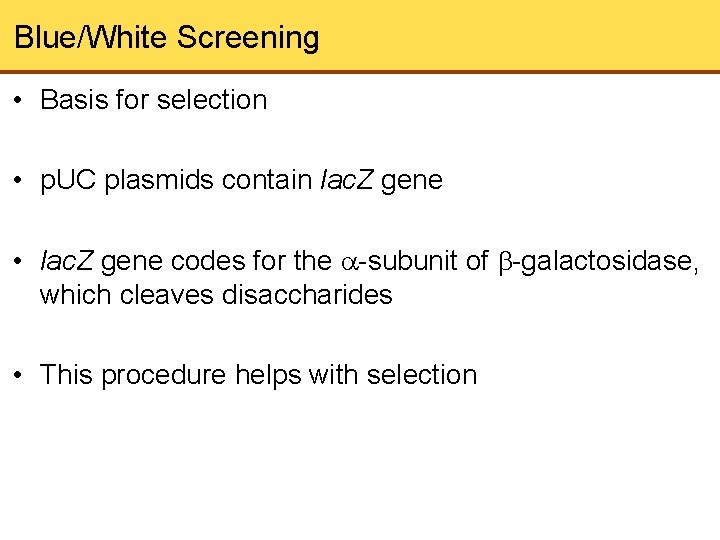
Blue/White Screening • Basis for selection • p. UC plasmids contain lac. Z gene • lac. Z gene codes for the -subunit of -galactosidase, which cleaves disaccharides • This procedure helps with selection
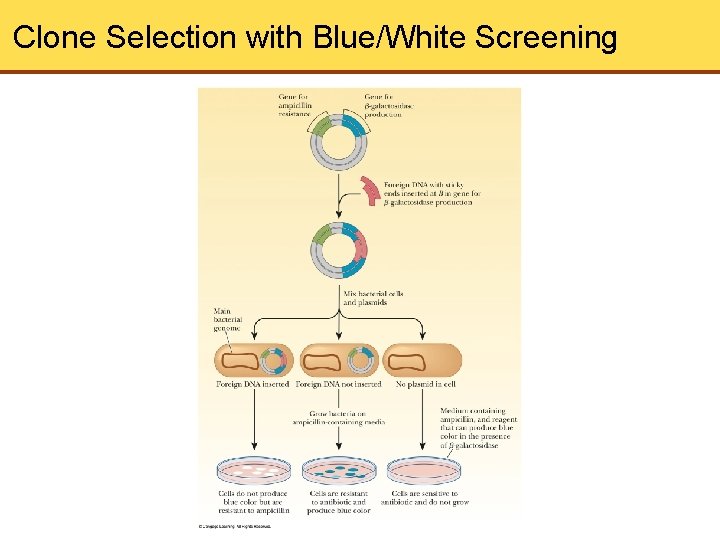
Clone Selection with Blue/White Screening

Cloning Summary • Cloning refers to creating identical populations • DNA can be combined by using restriction enzymes • The target DNA sequence is carried in some type of vector • The target DNA sequence is inserted into host organism • Organisms that carry the target DNA are identified through a process called selection
Genetic Engineering • When an organism is intentionally altered at the molecular level to exhibit different traits, it has been genetically engineered • One focus of genetic engineering has been gene therapy, where cells of specific tissues in a living person are altered in a way that alleviates the affects of a disease • DNA recombination can occur in nature • The reproductive power of bacteria can be used to express large quantities of a mammalian protein of interest, however, process can be complicated
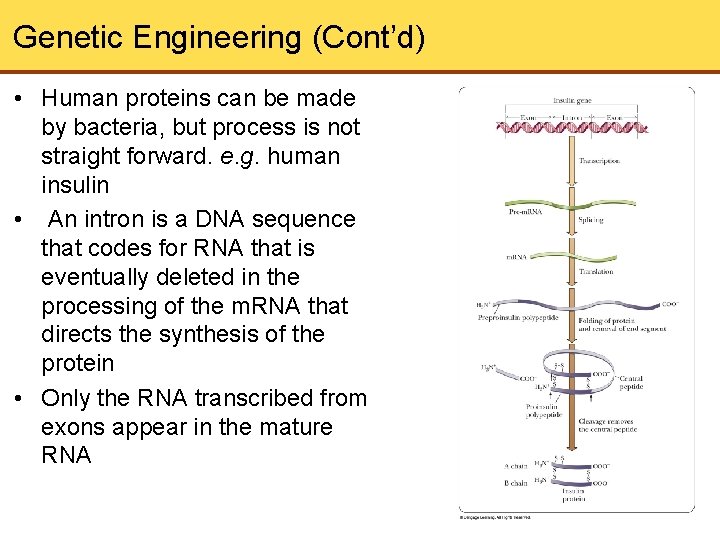
Genetic Engineering (Cont’d) • Human proteins can be made by bacteria, but process is not straight forward. e. g. human insulin • An intron is a DNA sequence that codes for RNA that is eventually deleted in the processing of the m. RNA that directs the synthesis of the protein • Only the RNA transcribed from exons appear in the mature RNA

Protein Expression Vectors • Plasmid vectors p. BR 322 and p. UC are cloning vectors • Vectors are used to insert foreign DNA and amplify it • If we want to produce protein from the foreign DNA, vectors are not good • Instead, expression vectors are used
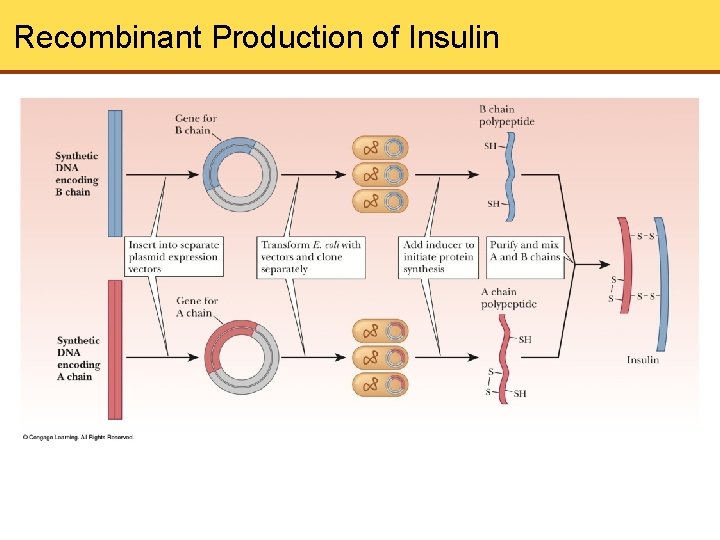
Recombinant Production of Insulin
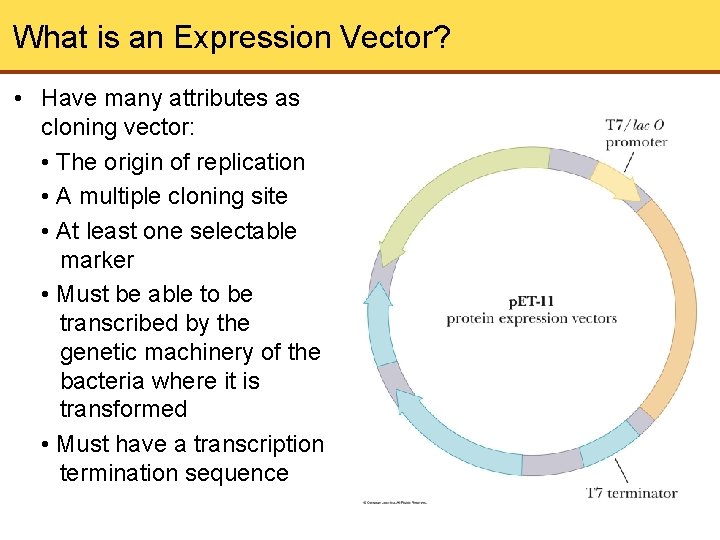
What is an Expression Vector? • Have many attributes as cloning vector: • The origin of replication • A multiple cloning site • At least one selectable marker • Must be able to be transcribed by the genetic machinery of the bacteria where it is transformed • Must have a transcription termination sequence
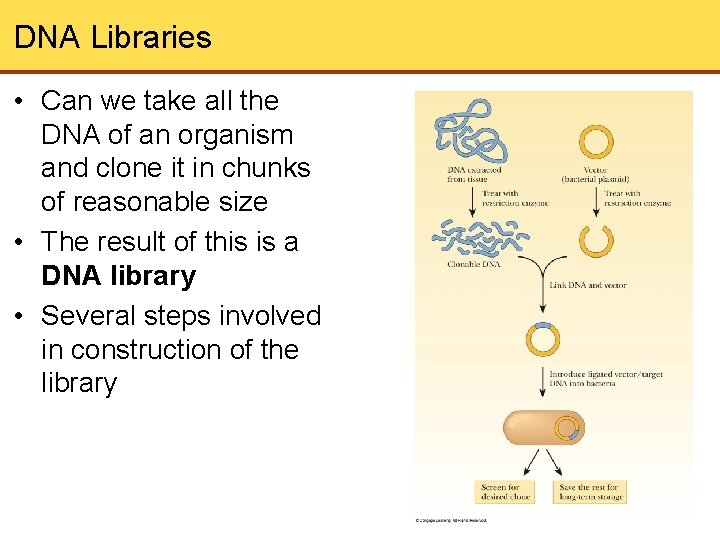
DNA Libraries • Can we take all the DNA of an organism and clone it in chunks of reasonable size • The result of this is a DNA library • Several steps involved in construction of the library
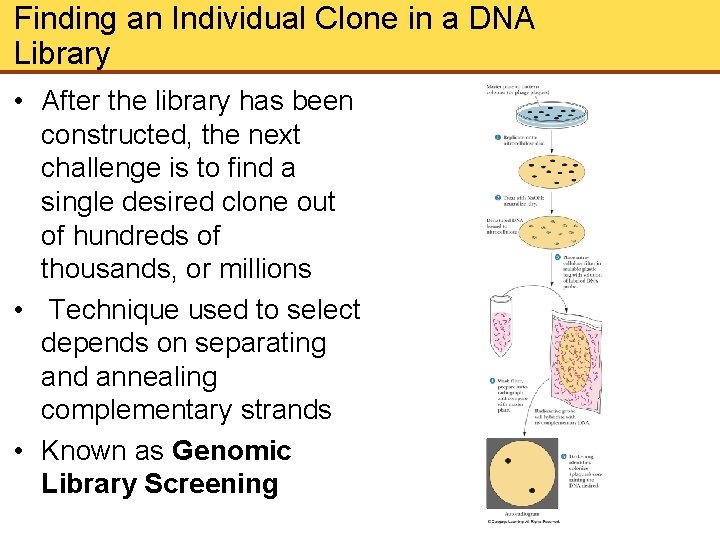
Finding an Individual Clone in a DNA Library • After the library has been constructed, the next challenge is to find a single desired clone out of hundreds of thousands, or millions • Technique used to select depends on separating and annealing complementary strands • Known as Genomic Library Screening
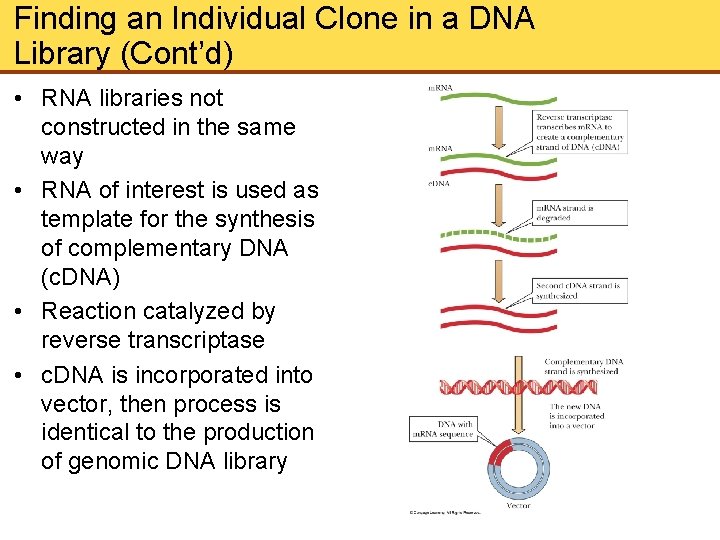
Finding an Individual Clone in a DNA Library (Cont’d) • RNA libraries not constructed in the same way • RNA of interest is used as template for the synthesis of complementary DNA (c. DNA) • Reaction catalyzed by reverse transcriptase • c. DNA is incorporated into vector, then process is identical to the production of genomic DNA library

Summary • A DNA library is a collection of clones of an entire genome • The genome is digested with restriction enzymes and the pieces are cloned into vectors, and transformed into cell lines • Specific radioactive probes to a sequence of interest are reacted to filters that have copies of the bacterial colonies in the library • A c. DNA library is constructed by using reverse transcriptase to make DNA from the m. RNA in a cell. This c. DNA is then used to construct a library similar to a genomic DNA library
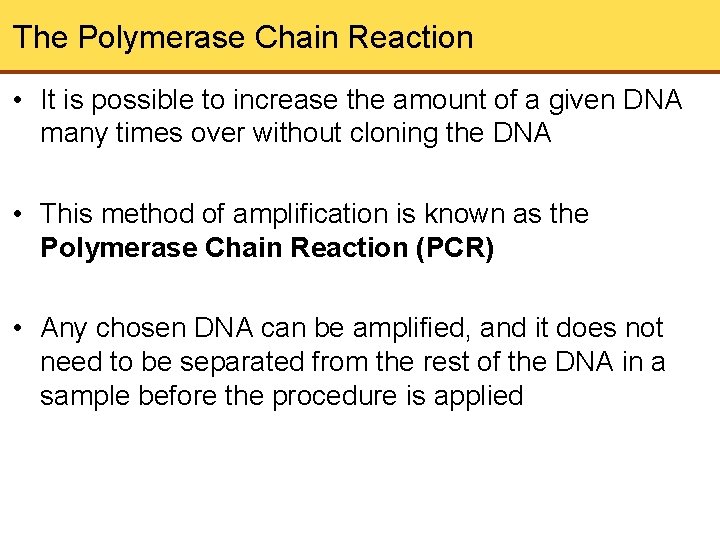
The Polymerase Chain Reaction • It is possible to increase the amount of a given DNA many times over without cloning the DNA • This method of amplification is known as the Polymerase Chain Reaction (PCR) • Any chosen DNA can be amplified, and it does not need to be separated from the rest of the DNA in a sample before the procedure is applied
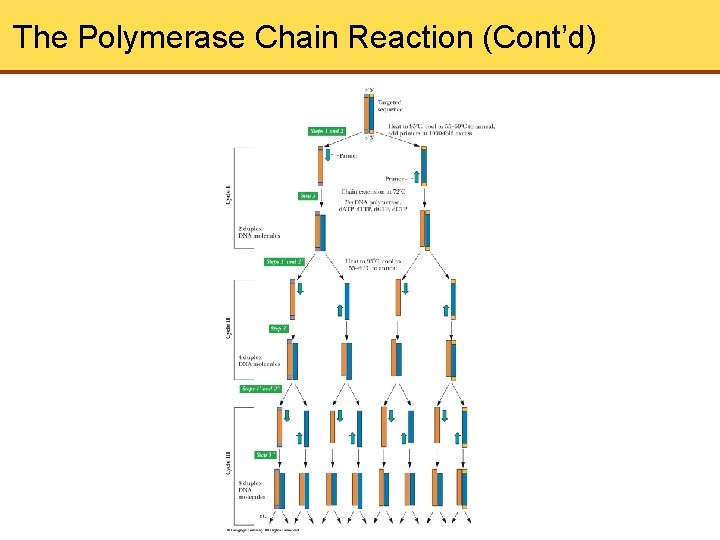
The Polymerase Chain Reaction (Cont’d)
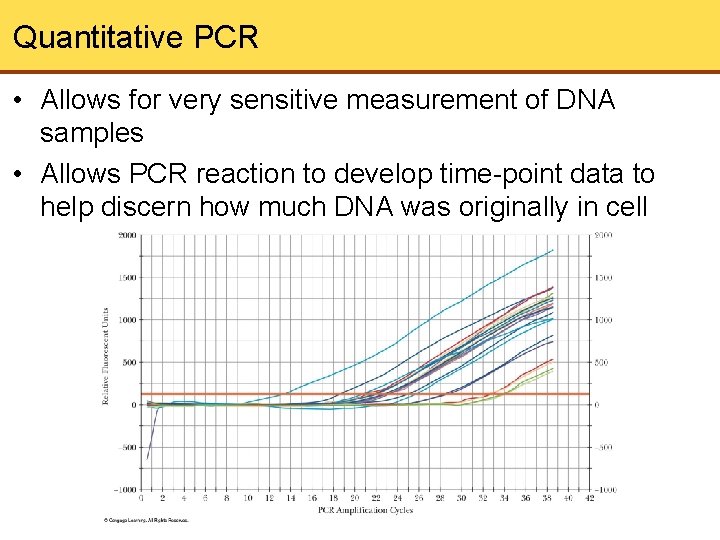
Quantitative PCR • Allows for very sensitive measurement of DNA samples • Allows PCR reaction to develop time-point data to help discern how much DNA was originally in cell

DNA Fingerprinting • DNA samples can be studied and compared by DNA fingerprinting • DNA is digested with restriction enzymes and then run on an agarose gel • When soaked in ethidium bromide, the DNA fragments can be seen directly under UV light • If greater sensitivity needed or if number of fragments would be too great to distinguish the bands, technique can be modified to show only selected DNA sequences • This begins with Southern blotting
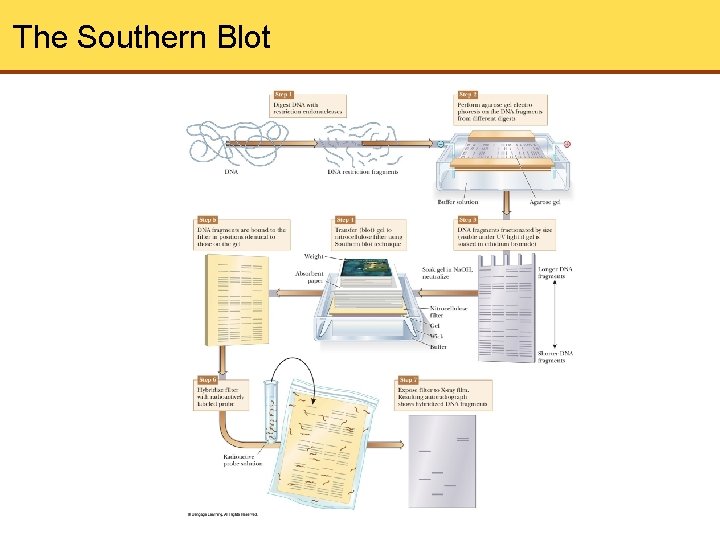
The Southern Blot
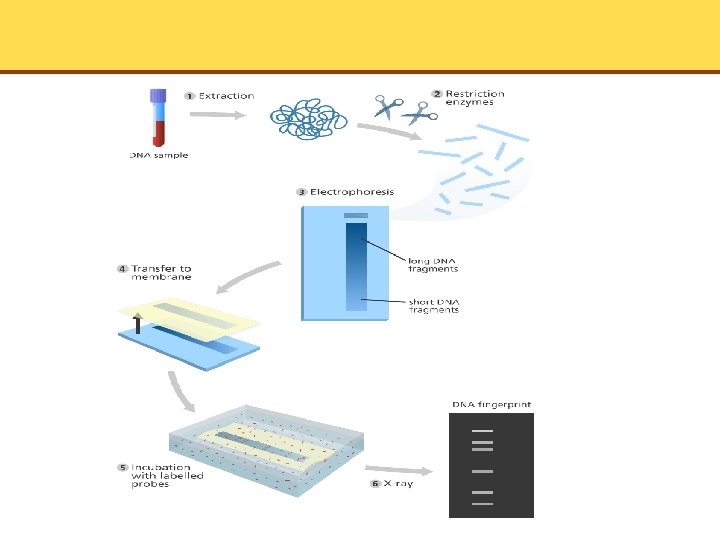
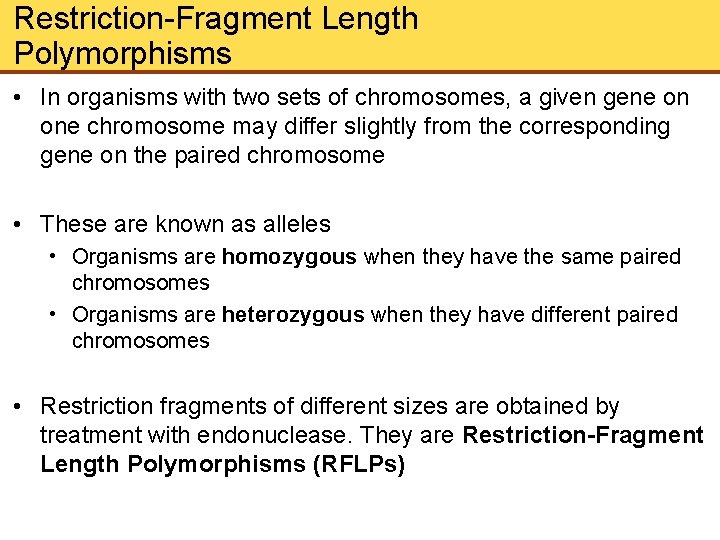
Restriction-Fragment Length Polymorphisms • In organisms with two sets of chromosomes, a given gene on one chromosome may differ slightly from the corresponding gene on the paired chromosome • These are known as alleles • Organisms are homozygous when they have the same paired chromosomes • Organisms are heterozygous when they have different paired chromosomes • Restriction fragments of different sizes are obtained by treatment with endonuclease. They are Restriction-Fragment Length Polymorphisms (RFLPs)
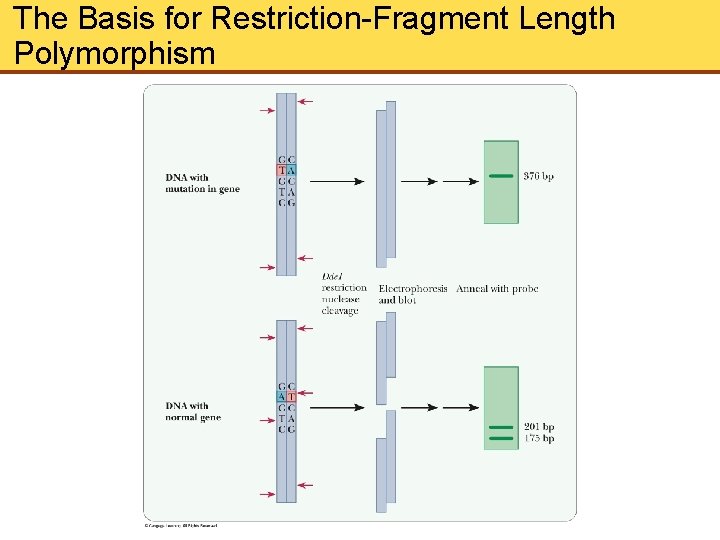
The Basis for Restriction-Fragment Length Polymorphism
Summary • A DNA fingerprint is created by digesting DNA with restriction enzymes, separating the pieces on a gel, and visualizing some of the pieces by using labeled probes • Differences in DNA patterns between different individuals are based on different base sequences of their DNA
Sequencing DNA • The nature and order of monomer units determine the properties of the whole molecule • The method devised by Sanger and Coulson for determining the base sequences of nucleic acids depends on selective interruption of oligonucleotide synthesis • A single-stranded DNA fragment whose sequence is to be determined is used as a template • The synthesis is interrupted at every possible site in the population of molecules depending on the presence of dd. NTPs
Sequencing DNA (Cont’d) • The incorporation of the dd. NTP into the growing chain causes termination at the point of incorporation • The DNA to be sequenced is mixed with a short oligonucleotide that serves as a primer for synthesis of the complementary strand • Gel electrophoresis is performed on each reaction mixture, and a band corresponding to each position of the chain termination appears • The sequence of the newly formed strand, complementary to the template DNA, can then be read from the sequencing gel
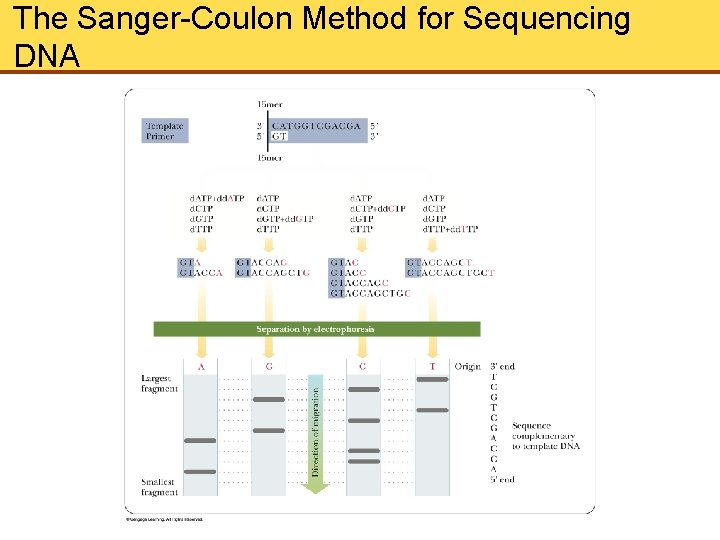
The Sanger-Coulon Method for Sequencing DNA
Summary • DNA can be sequenced by using several techniques, the most common being the chain termination method • Dideoxy nucleotides are used to terminate DNA synthesis. Multiple reactions are run with different dideoxy nucleotide in each reaction mix • The reactions produce a series of DNA fragments of different length that can be run on a gel and the sequence determined by tracking the different length fragments in the lanes with the four different dideoxy nucleotides
Genomics and Proteomics • Knowing the full DNA sequence of the human genome allows for the investigation for the causes of disease in a way that has not been possible until now • The proteome is a protein version of a genome • Proteomics is the study of interactions among all the proteins in a cell • Presently, due to genomics, we can study the “transcriptome” (all genes transcribed into RNA in the cell). Also “Metabolome”, and the “kinome. ”
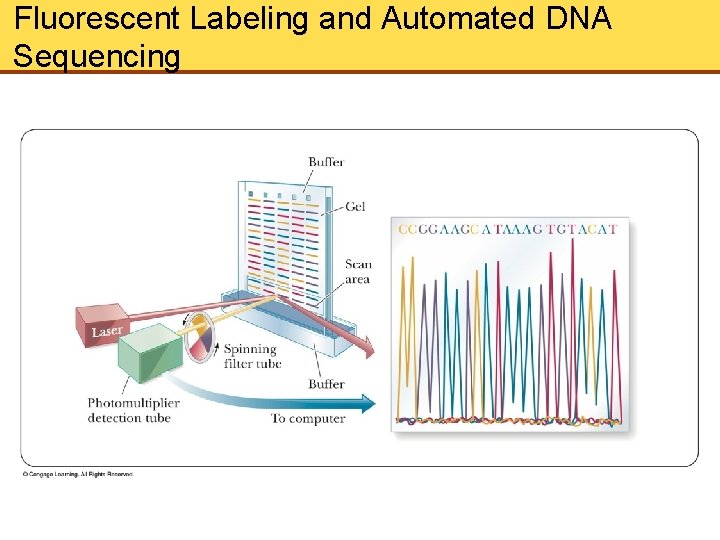
Fluorescent Labeling and Automated DNA Sequencing
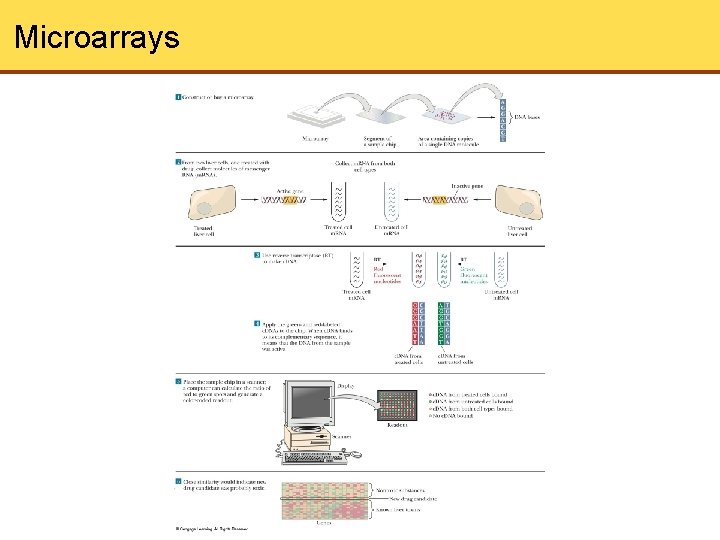
Microarrays
Summary • As more DNA sequences become available, it becomes possible to compare these sequences • Important medical applications are emerging, and new methods are making it possible to analyze large quantities of data • The proteome is the protein version of the genome. It refers to all of the proteins being expressed in a cell
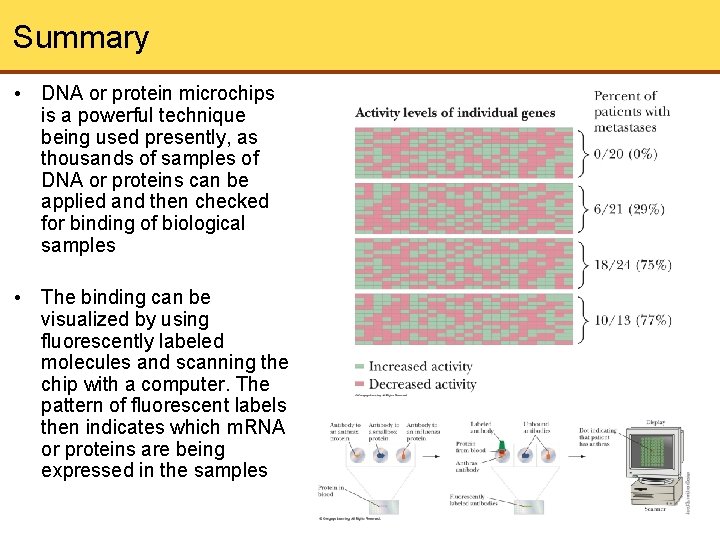
Summary • DNA or protein microchips is a powerful technique being used presently, as thousands of samples of DNA or proteins can be applied and then checked for binding of biological samples • The binding can be visualized by using fluorescently labeled molecules and scanning the chip with a computer. The pattern of fluorescent labels then indicates which m. RNA or proteins are being expressed in the samples
ASSIGNMENT • Cloning is the production of an organism genetically identical to another by means of nucleus transplantation. Why the practice of recombinant DNA technology and nucleus transplantation technology still dangerous? Justify your answer. • Cancer cell occurs with radiation and DNA-altered chemical during replication of DNA. How these situations occur? Justify your answer. • Centrifugal separation technique is common practice in clinical and research laboratory. What are the differences of preparative ultracentrifuges and analytical ultracentrifuge based on their principles? Justify your answer. • In genetic engineering, human insulin was produced in bacteria. What the mechanism of expression human insulin in bacteria? Justify your answer. • RIA and ELISA is an immunology study of the body immune responses. How the RIA and ELISA can be used as quantitative analysis of hormones, steroids and drugs? Justify your answer. • Study of new genome was related to DNA recombinant technology. How to identify of new genome? Justify your answer
- Slides: 48