Challenges of data management and analysis from 2
















- Slides: 44

Challenges of data management and analysis from 2 nd generation sequencing platforms October 10 2006 Please dial: +44 870 22 333 65 Pin: 444888 Copyright © 2004 Synamatix sdn bhd (538481 -U)

Challenges of data management and analysis from 2 nd generation sequencing platforms October 10 2006 Please dial: +44 870 22 333 65 Pin: 444888 Copyright © 2004 Synamatix sdn bhd (538481 -U)

Presenters Colin Hercus Zayed Albertyn 25 mer mapping 100 mer & polony mapping Copyright © 2006 Synamatix sdn bhd (538481 -U)

Introduction Copyright © 2006 Synamatix sdn bhd (538481 -U)

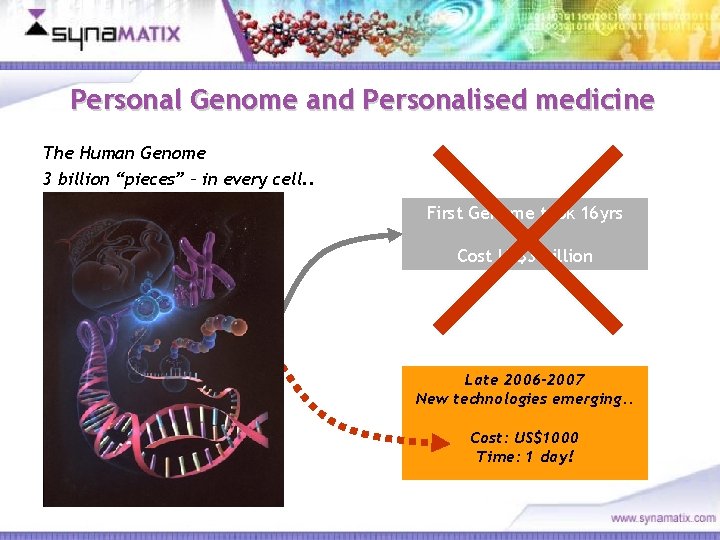
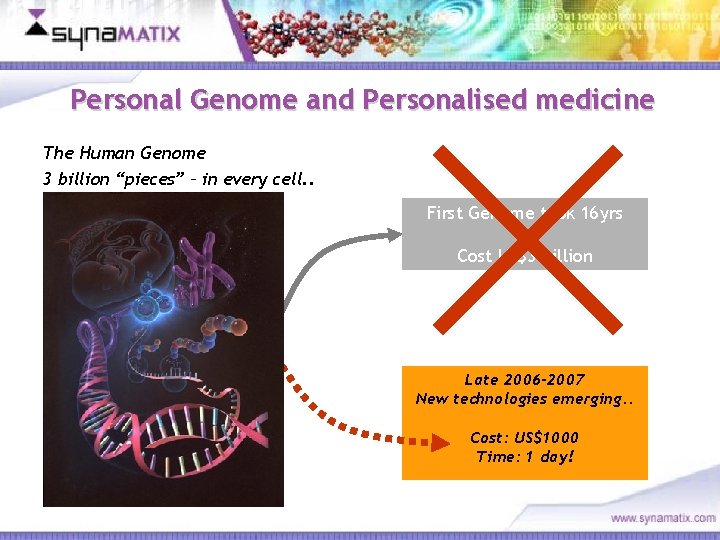
Personal Genome and Personalised medicine The Human Genome 3 billion “pieces” – in every cell. . First Genome took 16 yrs Cost US$3 billion Late 2006 -2007 New technologies emerging. . Cost: US$1000 Time: 1 day! Copyright © 2006 Synamatix sdn bhd (538481 -U)

Personal Genome and Personalised medicine The Human Genome 3 billion “pieces” – in every cell. . First Genome took 16 yrs Cost US$3 billion Late 2006 -2007 New technologies emerging. . Cost: US$1000 Time: 1 day! Copyright © 2006 Synamatix sdn bhd (538481 -U)

Variety of approaches towards ULCS Copyright © 2006 Synamatix sdn bhd (538481 -U)

Methods Copyright © 2006 Synamatix sdn bhd (538481 -U)

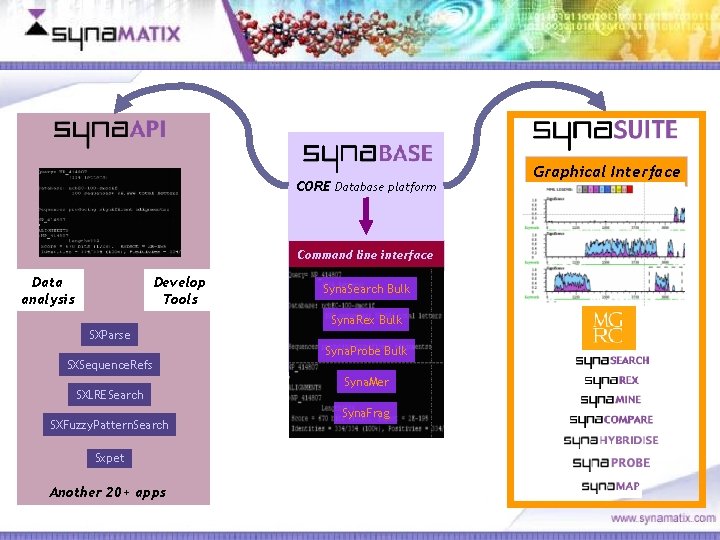
CORE Database platform Command line interface Data analysis Develop Tools Syna. Search Bulk Syna. Rex Bulk SXParse Syna. Probe Bulk SXSequence. Refs SXLRESearch SXFuzzy. Pattern. Search Syna. Mer Syna. Frag Sxpet Another 20+ apps Copyright © 2006 Synamatix sdn bhd (538481 -U) Graphical Interface

How? Copyright © 2006 Synamatix sdn bhd (538481 -U)

What do we know about data ? Similarity & association Common PATTERNS and functionality Copyright © 2006 Synamatix sdn bhd (538481 -U)

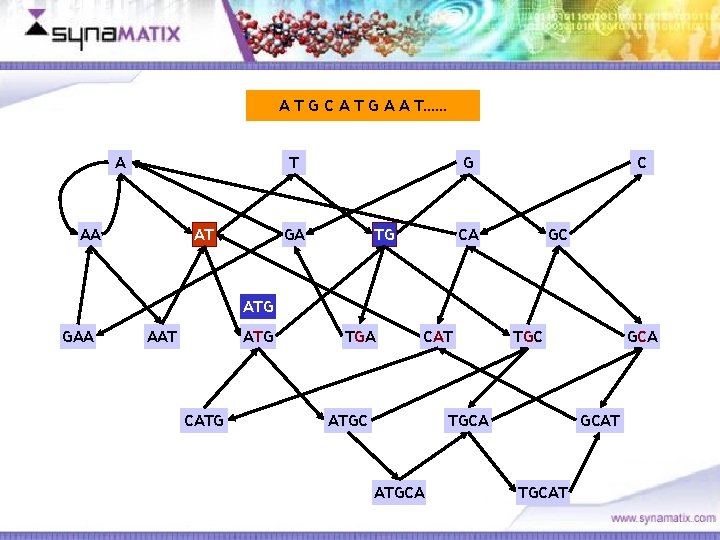
A T G C A T G A A T…… A T AA AT G GA TG C CA GC ATG GAA AAT ATG CATG TGA CAT ATGC TGCA ATGCA Copyright © 2006 Synamatix sdn bhd (538481 -U) GCAT TGCAT

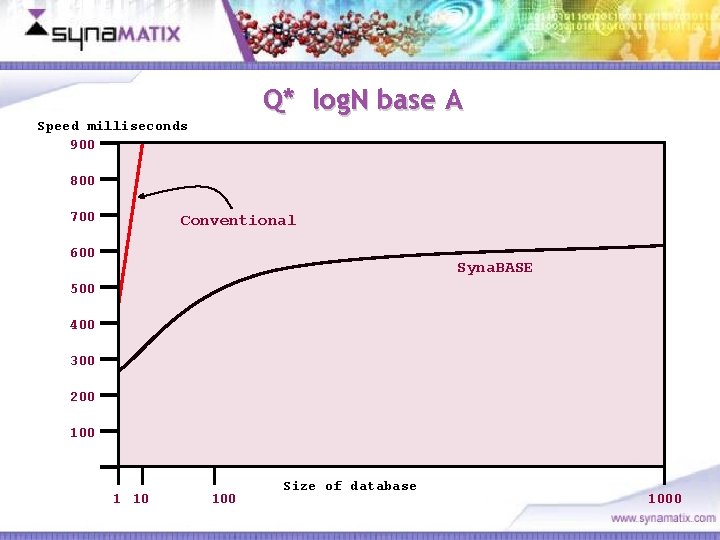
Q* log. N base A Speed milliseconds 900 800 700 Conventional 600 Syna. BASE 500 400 300 200 1 10 100 Size of database Copyright © 2006 Synamatix sdn bhd (538481 -U) 1000

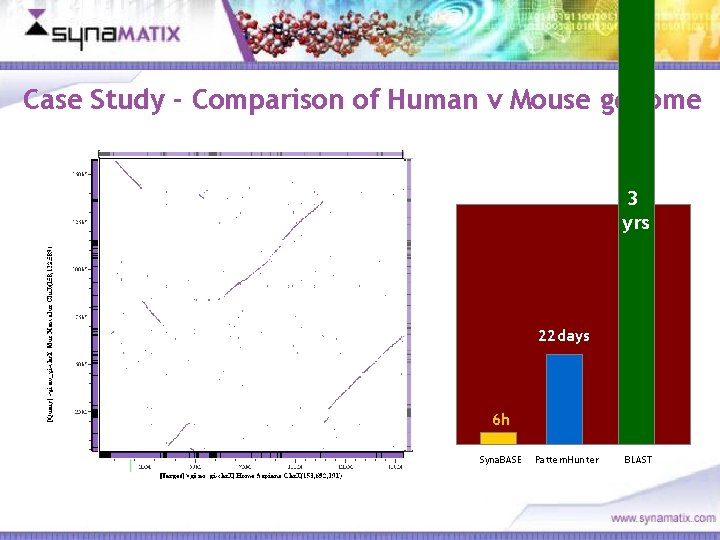
Case Study - Comparison of Human v Mouse genome 3 yrs 22 days 6 h Syna. BASE Copyright © 2006 Synamatix sdn bhd (538481 -U) Pattern. Hunter BLAST

Results Copyright © 2006 Synamatix sdn bhd (538481 -U)

Read mapping Variety of novel methods for genome sequencing Shorter reads with higher coverage 25 mers - Solexa 100 -200 mers – 454 Polony reads Larger volumes of sequence data Error rates much higher than Sanger method Computationally Intractable for conventional bioinformatics applications Copyright © 2006 Synamatix sdn bhd (538481 -U)

Mapping 25 mers Copyright © 2006 Synamatix sdn bhd (538481 -U)

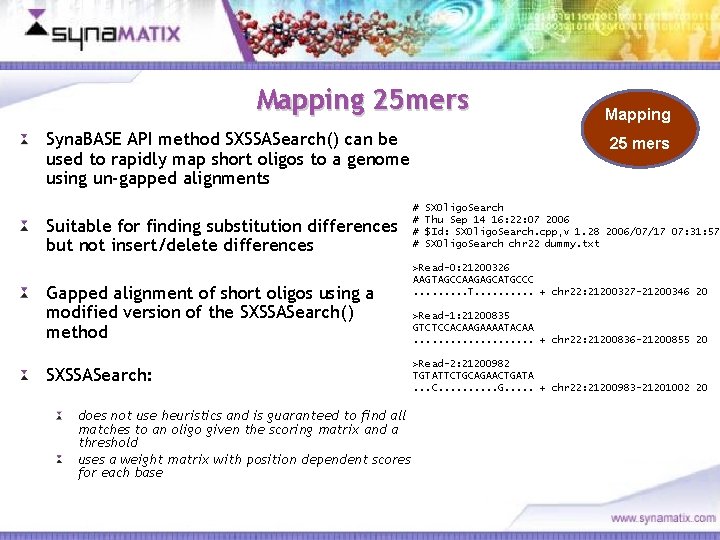
Mapping 25 mers Syna. BASE API method SXSSASearch() can be used to rapidly map short oligos to a genome using un-gapped alignments Suitable for finding substitution differences but not insert/delete differences Gapped alignment of short oligos using a modified version of the SXSSASearch() method Mapping 25 mers # # SXOligo. Search Thu Sep 14 16: 22: 07 2006 $Id: SXOligo. Search. cpp, v 1. 28 2006/07/17 07: 31: 57 SXOligo. Search chr 22 dummy. txt >Read-0: 21200326 AAGTAGCCAAGAGCATGCCC. . T. . + chr 22: 21200327 -21200346 20 >Read-1: 21200835 GTCTCCACAAGAAAATACAA. . . . . + chr 22: 21200836 -21200855 20 >Read-2: 21200982 TGTATTCTGCAGAACTGATA. . . C. . G. . . + chr 22: 21200983 -21201002 20 SXSSASearch: does not use heuristics and is guaranteed to find all matches to an oligo given the scoring matrix and a threshold uses a weight matrix with position dependent scores for each base Copyright © 2006 Synamatix sdn bhd (538481 -U)

Very fast and flexible approach Mapping 25 mers Example: 350, 000 reads can be mapped in 125 sec - 3 per ms Makes approach suitable for reads that have varying quality over their length Mismatch penalty can be reduced towards the 3’ end of reads Quality or Probability of being correct 1. 0 0 25 Copyright © 2006 Synamatix sdn bhd (538481 -U)

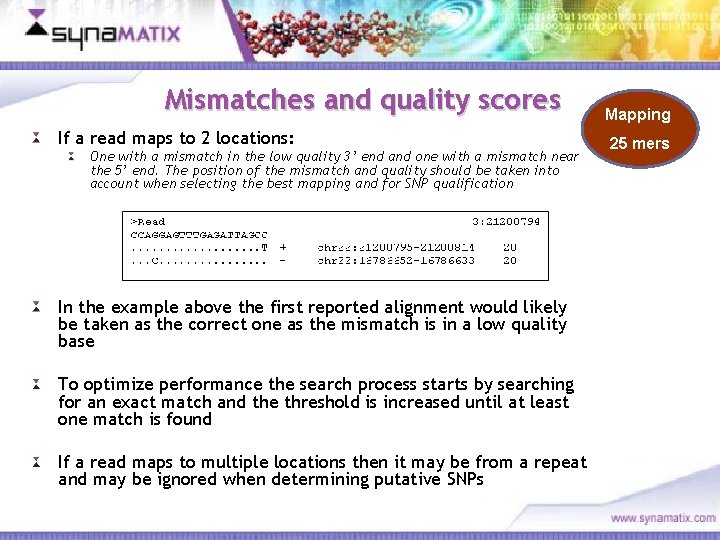
Mismatches and quality scores If a read maps to 2 locations: One with a mismatch in the low quality 3’ end and one with a mismatch near the 5’ end. The position of the mismatch and quality should be taken into account when selecting the best mapping and for SNP qualification In the example above the first reported alignment would likely be taken as the correct one as the mismatch is in a low quality base To optimize performance the search process starts by searching for an exact match and the threshold is increased until at least one match is found If a read maps to multiple locations then it may be from a repeat and may be ignored when determining putative SNPs Copyright © 2006 Synamatix sdn bhd (538481 -U) Mapping 25 mers

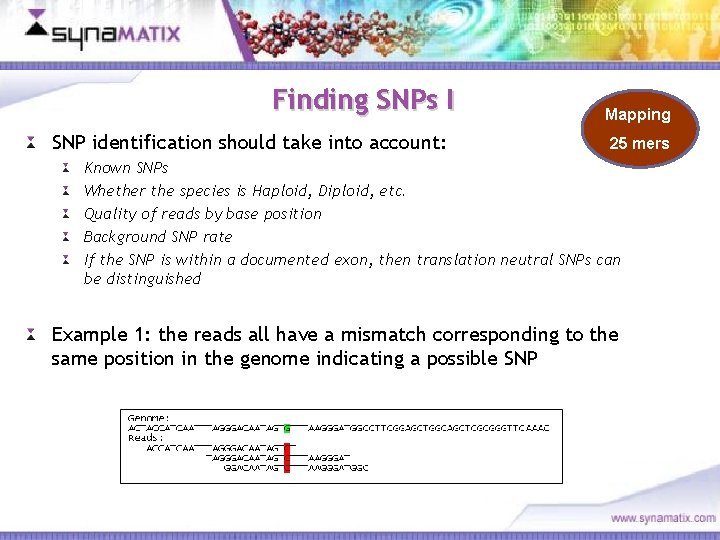
Finding SNPs I SNP identification should take into account: Mapping 25 mers Known SNPs Whether the species is Haploid, Diploid, etc. Quality of reads by base position Background SNP rate If the SNP is within a documented exon, then translation neutral SNPs can be distinguished Example 1: the reads all have a mismatch corresponding to the same position in the genome indicating a possible SNP Copyright © 2006 Synamatix sdn bhd (538481 -U)

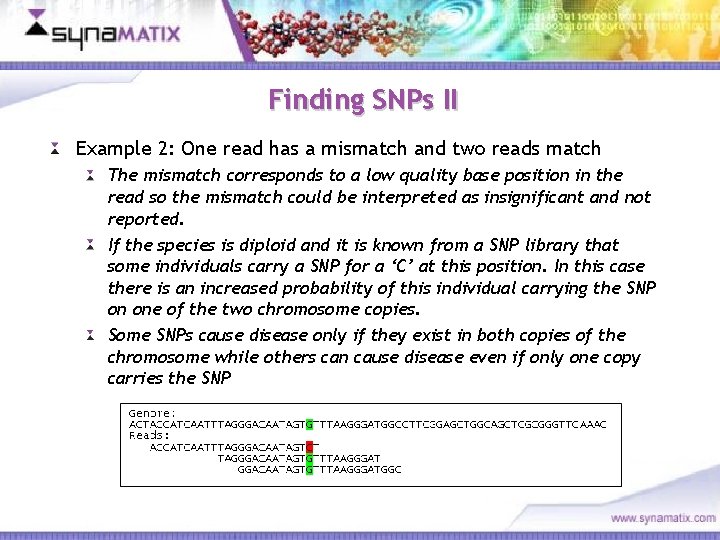
Finding SNPs II Example 2: One read has a mismatch and two reads match The mismatch corresponds to a low quality base position in the read so the mismatch could be interpreted as insignificant and not reported. If the species is diploid and it is known from a SNP library that some individuals carry a SNP for a ‘C’ at this position. In this case there is an increased probability of this individual carrying the SNP on one of the two chromosome copies. Some SNPs cause disease only if they exist in both copies of the chromosome while others can cause disease even if only one copy carries the SNP Copyright © 2006 Synamatix sdn bhd (538481 -U)

Summary Mapping 25 mers Mapping of short reads achieved at very high throughput – less than 1 ms Position specific scoring allows variable quality reads to be mapped Statistical analysis of mismatches to qualify SNPs Copyright © 2006 Synamatix sdn bhd (538481 -U)

Mapping 100 mers Copyright © 2006 Synamatix sdn bhd (538481 -U)

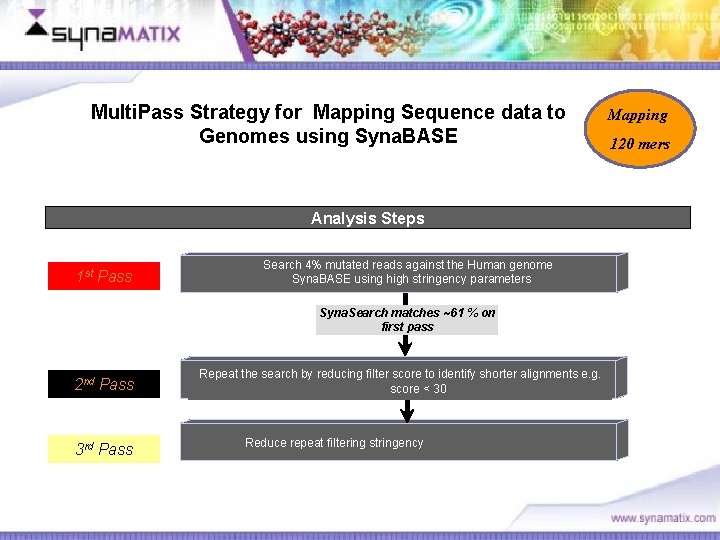
Multi. Pass Strategy for Mapping Sequence data to Genomes using Syna. BASE Analysis Steps 1 st Pass Search 4% mutated reads against the Human genome Syna. BASE using high stringency parameters Syna. Search matches ~61 % on first pass 2 nd Pass 3 rd Pass Repeat the search by reducing filter score to identify shorter alignments e. g. score < 30 Reduce repeat filtering stringency Copyright © 2006 Synamatix sdn bhd (538481 -U) Mapping 120 mers

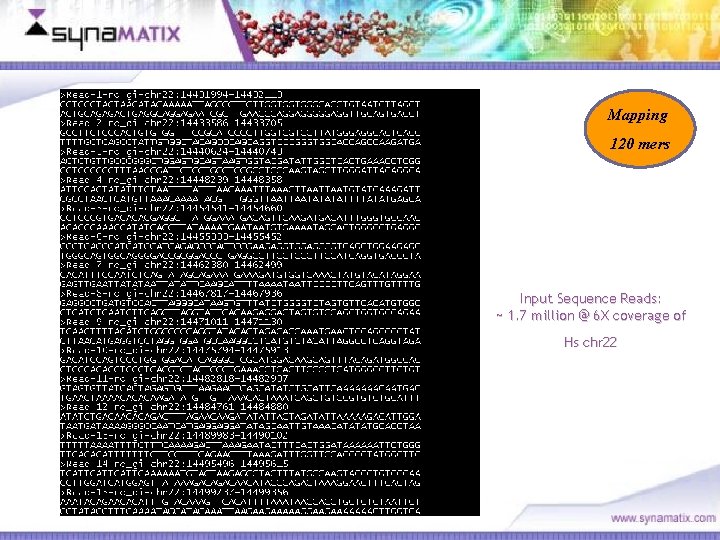
Mapping 120 mers Input Sequence Reads: ~ 1. 7 million @ 6 X coverage of Hs chr 22 Copyright © 2006 Synamatix sdn bhd (538481 -U)

Mapping 120 mers Copyright © 2006 Synamatix sdn bhd (538481 -U)

Analysis of Results Mapping 120 mers View read placement along chromosome Calculate mapping efficiency 1. 7 m reads mapped to human genome in 53 min 22 seconds Copyright © 2006 Synamatix sdn bhd (538481 -U)

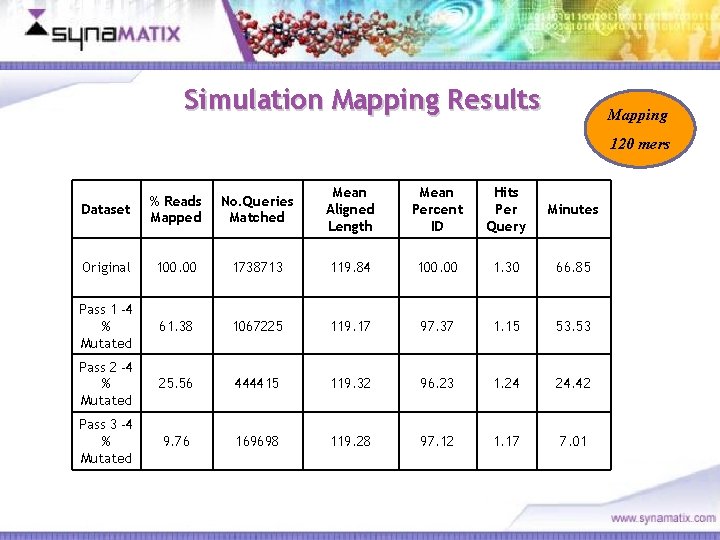
Simulation Mapping Results Mapping 120 mers Dataset % Reads Mapped No. Queries Matched Mean Aligned Length Mean Percent ID Hits Per Query Minutes Original 100. 00 1738713 119. 84 100. 00 1. 30 66. 85 Pass 1 -4 % Mutated 61. 38 1067225 119. 17 97. 37 1. 15 53. 53 Pass 2 -4 % Mutated 25. 56 444415 119. 32 96. 23 1. 24 24. 42 Pass 3 -4 % Mutated 9. 76 169698 119. 28 97. 12 1. 17 7. 01 Copyright © 2006 Synamatix sdn bhd (538481 -U)

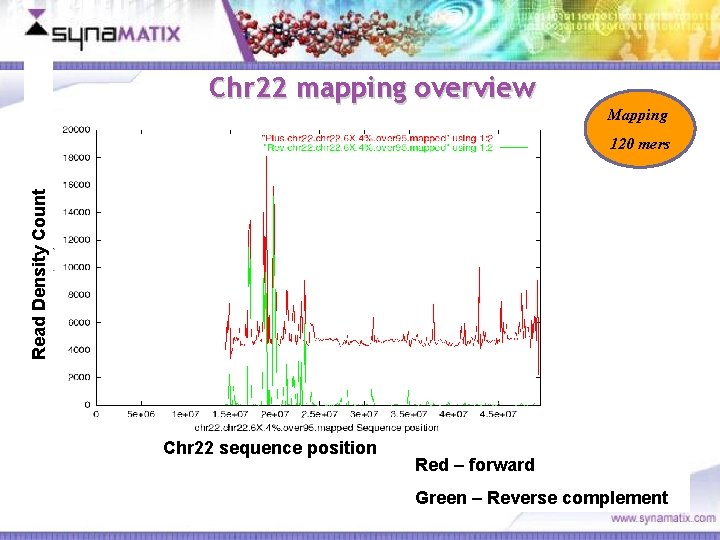
Chr 22 mapping overview Mapping Read Density Count 120 mers Chr 22 sequence position Red – forward Green – Reverse complement Copyright © 2006 Synamatix sdn bhd (538481 -U)

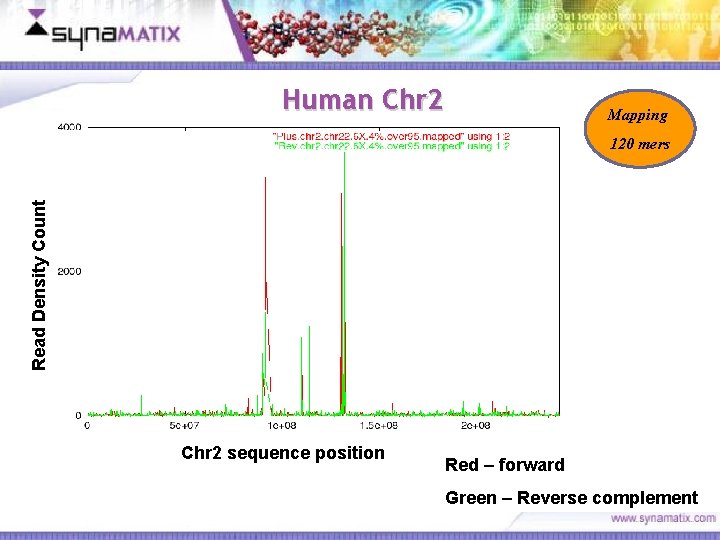
Human Chr 2 Mapping Read Density Count 120 mers Chr 2 sequence position Red – forward Green – Reverse complement Copyright © 2006 Synamatix sdn bhd (538481 -U)

Viewing Results Mapping 120 mers Gbrowse: Community-based system to view results Numerous customisations to show sequence coverage Analyze read mappings in the context of Known genes Repeats and variations (SNP) Comparative genomics Copyright © 2006 Synamatix sdn bhd (538481 -U)

Mapping 120 mers Copyright © 2006 Synamatix sdn bhd (538481 -U)

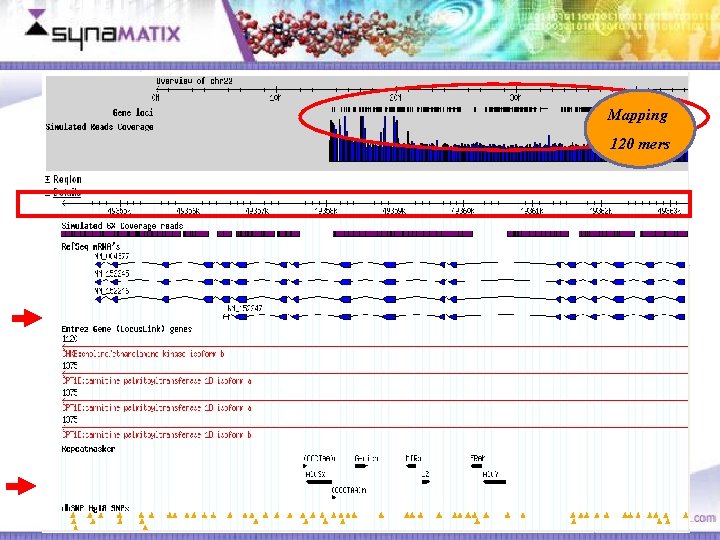
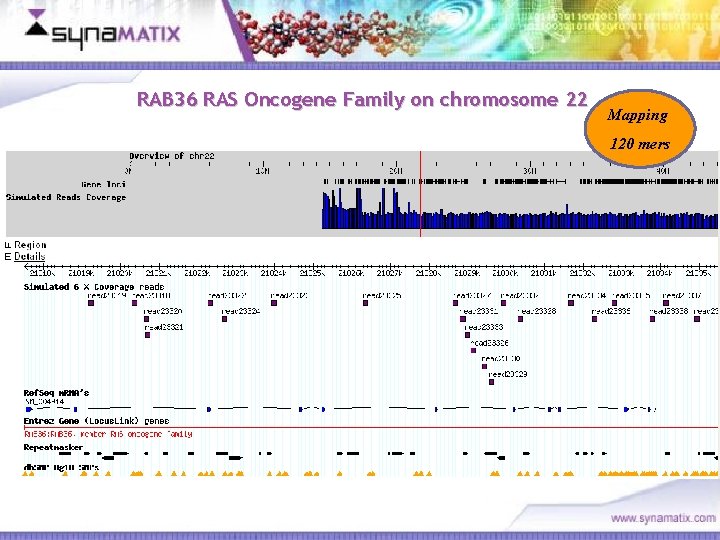
RAB 36 RAS Oncogene Family on chromosome 22 Mapping 120 mers Copyright © 2006 Synamatix sdn bhd (538481 -U)

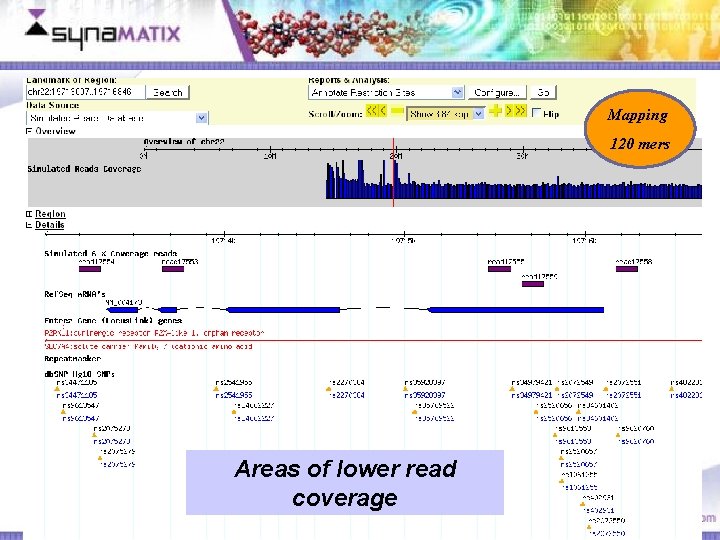
Mapping 120 mers Areas of lower read coverage Copyright © 2006 Synamatix sdn bhd (538481 -U)

Conclusions Mapping 120 mers Very significant performance improvements compared to Mega. BLAST – <100 ms per read Very high coverage attained by using multi-pass strategy Over 95% coverage Remaining 5% are repeats High specificity – less matches per read Enables multiple human genomes to be processed per day Copyright © 2006 Synamatix sdn bhd (538481 -U)

Mapping Polony reads 5 mers Copyright © 2006 Synamatix sdn bhd (538481 -U)

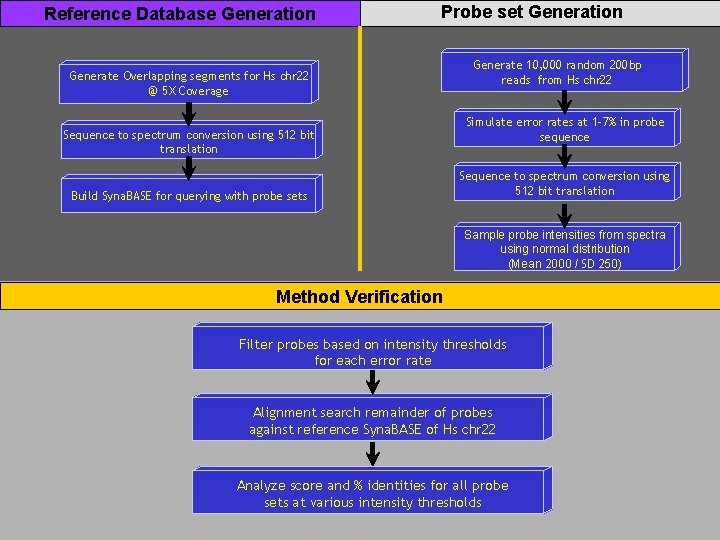
Polony sequencing read mapping 5 mers polony reads Convert genomic sequences to spectra Sample random probe sets from random chromosomal regions Filter probe sets using probe intensity spectra Query probe sets against genome database Copyright © 2006 Synamatix sdn bhd (538481 -U)

Reference Database Generation Probe set Generation Generate Overlapping segments for Hs chr 22 @ 5 X Coverage Sequence to spectrum conversion using 512 bit translation Build Syna. BASE for querying with probe sets Generate 10, 000 random 200 bp reads from Hs chr 22 Simulate error rates at 1 -7% in probe sequence Sequence to spectrum conversion using 512 bit translation Sample probe intensities from spectra using normal distribution (Mean 2000 / SD 250) Method Verification Filter probes based on intensity thresholds for each error rate Alignment search remainder of probes against reference Syna. BASE of Hs chr 22 Analyze score and % identities for all probe sets at various intensity thresholds Copyright © 2006 Synamatix sdn bhd (538481 -U)

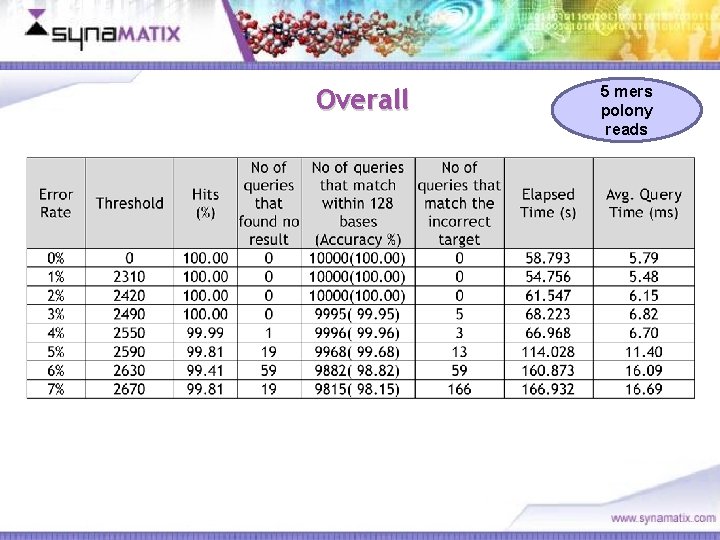
Overall Copyright © 2006 Synamatix sdn bhd (538481 -U) 5 mers polony reads

Advantages 5 mers polony reads Time taken to conduct 0% to 4% searches – around 6 ms Enhanced performance to the Syna. BASE engine & associated algorithms 100% hits matched for 1 -3 % error margin data ~15 million searches against a reference genome in 1 day Copyright © 2006 Synamatix sdn bhd (538481 -U)

Conclusion Copyright © 2006 Synamatix sdn bhd (538481 -U)

Summary Syna. BASE used as database PLATFORM Unique, leads to massive increases in speed and scalability Applied to the 3 main classes of reads from 2 nd generation sequencing platforms 100 s of fold faster than conventional approaches Specificity and accuracy enhanced due to exhaustive nature of Syna. BASE Copyright © 2006 Synamatix sdn bhd (538481 -U)

Thank you Please email questions to: enquiries@synamatix. com Copyright © 2006 Synamatix sdn bhd (538481 -U)