Challenges in data management and running DNA sequencing




















- Slides: 24

Challenges in data management and running DNA sequencing experiments on grid EGI community forum – March 27, 2012 Barbera van Schaik, Mark Santcroos, Vladimir Korkhov, Aldo Jongejan, Marcel Willemsen, Antoine van Kampen and Silvia Olabarriaga

Sequence facility Introduction to the groups Bioinformatics NGS team Research laboratories e-Bio. Science team grid

Presented at EGEE 2010: BLAST for virus discovery Proof of concept: 30 x speed-up Application is currently used by the virus discovery unit “Last week we did a new sequence run and we found 3 new viruses the next day!”

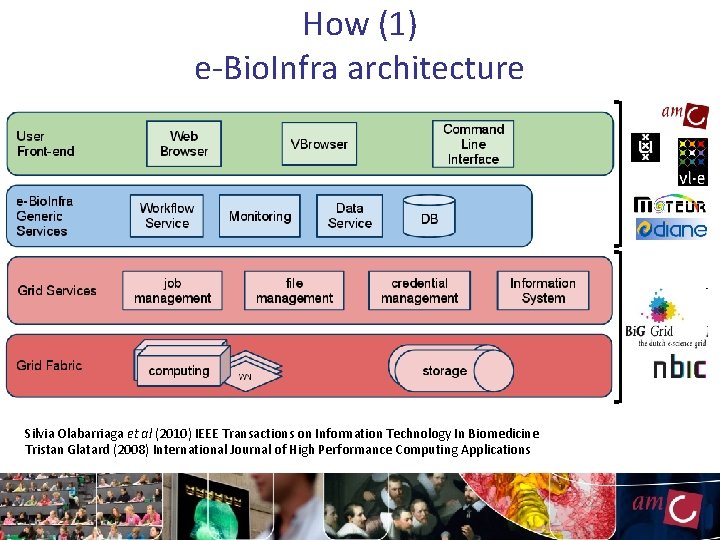
How (1) e-Bio. Infra architecture Silvia Olabarriaga et al (2010) IEEE Transactions on Information Technology In Biomedicine Tristan Glatard (2008) International Journal of High Performance Computing Applications

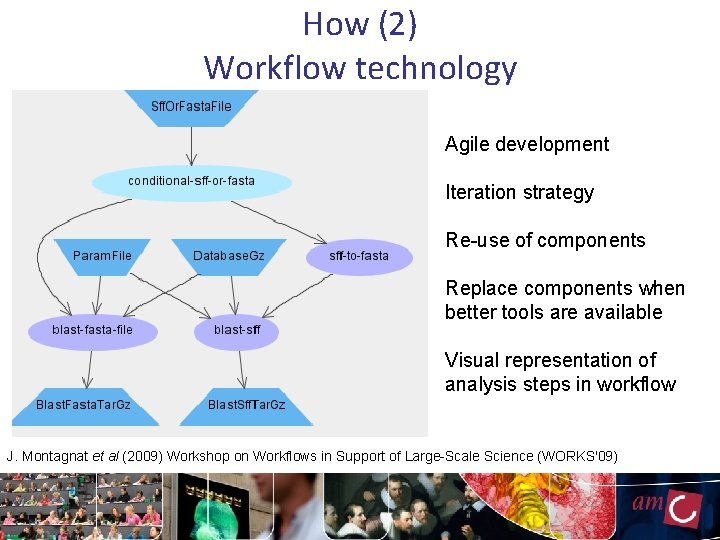
How (2) Workflow technology Agile development Iteration strategy Re-use of components Replace components when better tools are available Visual representation of analysis steps in workflow J. Montagnat et al (2009) Workshop on Workflows in Support of Large-Scale Science (WORKS'09)

Changes: diversity of analyses Which gene(s) cause disease Z? We have sequenced 20 bacterial genomes, what are the commonalities / differences? Are there specific micro. RNAs in HIV infected patients? Which genes are differentially expressed in situation X versus Y? Workflows have been implemented for these cases

Common in most projects: BWA • Aligns sequences to a reference database – Human genome – HIV genome – Bacterial genome • Especially designed for shorter sequences • Puts entire database in memory and aligns all experiment sequences • Run time almost linear to the amount of sequences http: //bio-bwa. sourceforge. net/

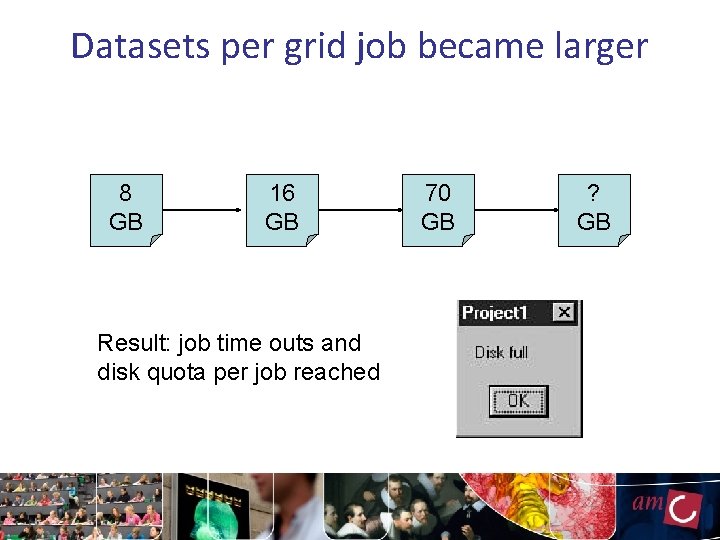
Changes: expansion of the DNA sequence facility ~1 GB per run ~120 ~60 GB GBper perrun ~120 GB per run In total around 16 TB per year After data analysis: 10 x size of the input data

Datasets per grid job became larger 8 GB 16 GB Result: job time outs and disk quota per job reached 70 GB ? GB

Improvements for BWA – split the input data + speed-up + smaller files per job Split Merge more jobs → more failed jobs

Check Implemented loops in workflow Checks if all files are generated Split Process http: //www. bioinformaticslaboratory. nl/twiki/bin/view/EBio. Science/Creating. Workflows

More changes and challenges: analyzing many big datasets Total raw data: 45 TB After alignment: 10 x increase Project partners are performing consecutive analyses on grid http: //www. dutchgenomeproject. com/

But first… getting the data on grid storage • This step less than ideal – It took one week to transfer 10 TB • Luckily there is a more efficient system now fers s n a r t f o e These typ ge) a r o t s d i r (HD > g ur c c o y l e t i will defin n more ofte

After data analysis: share results WIKI LFC http: //www. beehub. nl/ Tomorrow 11: 20, Tom Visser (Sara), this room

Changes in the workflow engine 2 - Needed to convert all component descriptions and workflows + End-users from Virus Discovery didn’t notice (except changes web-service URL and monitoring dashboard)

New changes ahead Bioinformaticians just got introduced to the portal Need to convert all 150 applications (again) ?

Why go through all this trouble? Why not write scripts in stead of workflows? Why not buy a bigger cluster?

Tools for next generation sequencing 500 new tools for sequencing in the past two years! http: //seqanswers. com/wiki/Software Better method available? Just replace component.

And … more data is expected Data throughput for each DNA sequence method http: //www. wellcome. ac. uk/Education-resources/Teaching-and-education/Big-Picture/All-issues/Genes-Genomes-and-Health/WTDV 027167. htm

Genome projects • • • Human genome project (1 individual) Exome sequencing (~10 individuals) Genome of the Netherlands (770 individuals) 1000 genome project (1000 individuals) UK 10 K project (10, 000 individuals) … URLs are in notes of this presentation

Finally: An example of an in-house project Measure gene activity Measure non-proteincoding gene activity Search for mutations causing disease (exome sequencing)

Verification of de novo mutations • De novo mutations found in Nicolaides Baraitser patients • Reviewers: Are these mutations specific for the disease? • Deadline: yesterday : ) Implementation workflow and gather input data: 2 weeks Variants of 223 healthy people Variants of 770 healthy people Run time: 1 day Annotation of variants Repeat with more samples Run time: 1. 5 day

How e-science changes the work for bioinformaticians and biomedical reachers • • Respond to requests quickly Share both data and methods Analyze multiple datasets at once Work on several projects simultaneously

Acknowledgements Virus discovery unit, AMC Lia van der Hoek Bas Oude Munnink Michel de Vries Department of genome analysis, AMC Frank Baas Ted Bradley Marja Jakobs Department of Pediatrics, AMC Raoul Hennekam University of Amsterdam Piter de Boer Bioinformatics Laboratory, AMC Antoine van Kampen Bi. G Grid Jan Just Keijser Tom Visser Grid support NGS bioinformatics team Aldo Jongejan Marcel Willemsen Modalis, France Johan Montagnat Creatis, France Tristan Glatard e-Bioscience team Silvia Olabarriaga Mark Santcroos Vladimir Korkhov Souley Madougou Kyriacos Neocleous Shayan Shahand Laboratory division of AMC http: //www. bioinformaticslaboratory. nl/