CH 339 K Proteins Primary Structure Purification and

















































- Slides: 145

CH 339 K Proteins: Primary Structure, Purification, and Sequencing

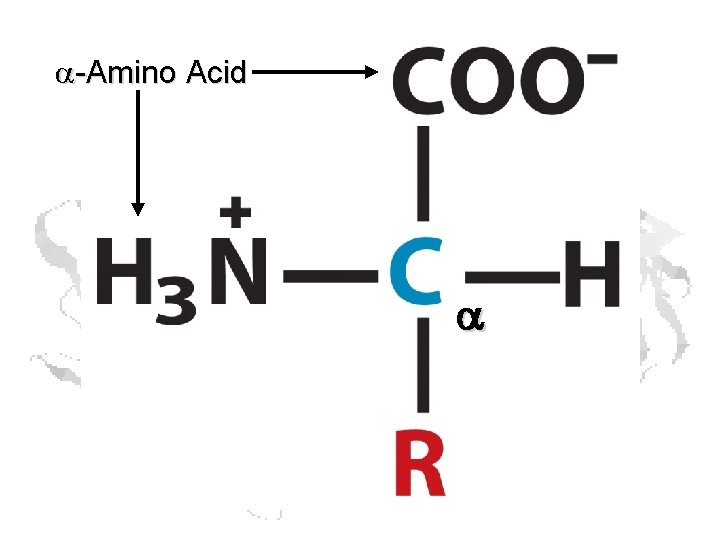
a-Amino Acid a

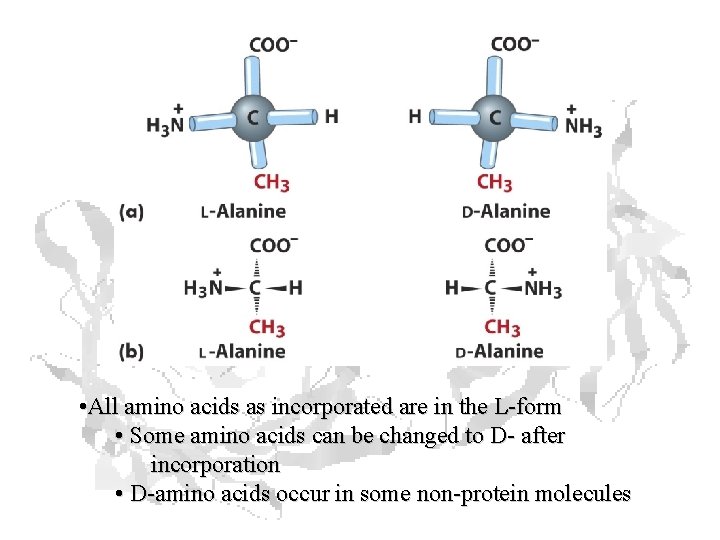
• All amino acids as incorporated are in the L-form • Some amino acids can be changed to D- after incorporation • D-amino acids occur in some non-protein molecules

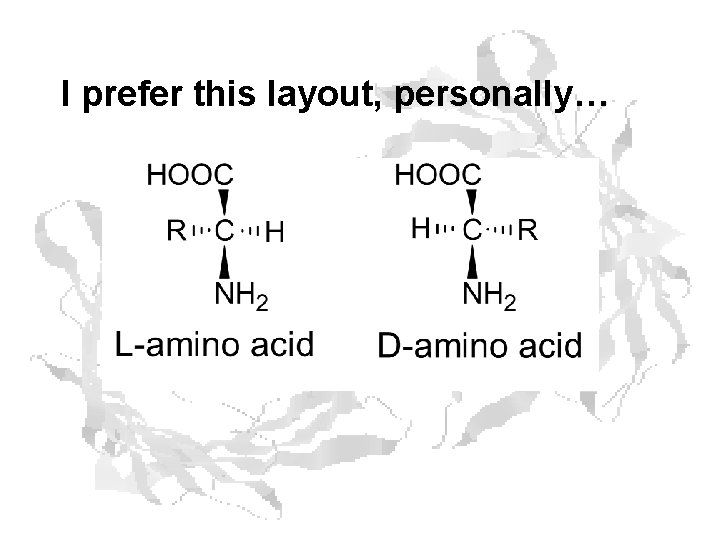
I prefer this layout, personally…



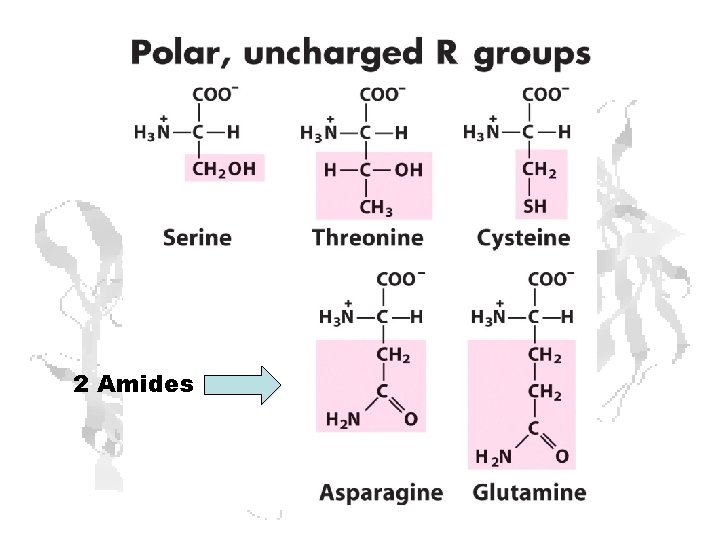
2 Amides


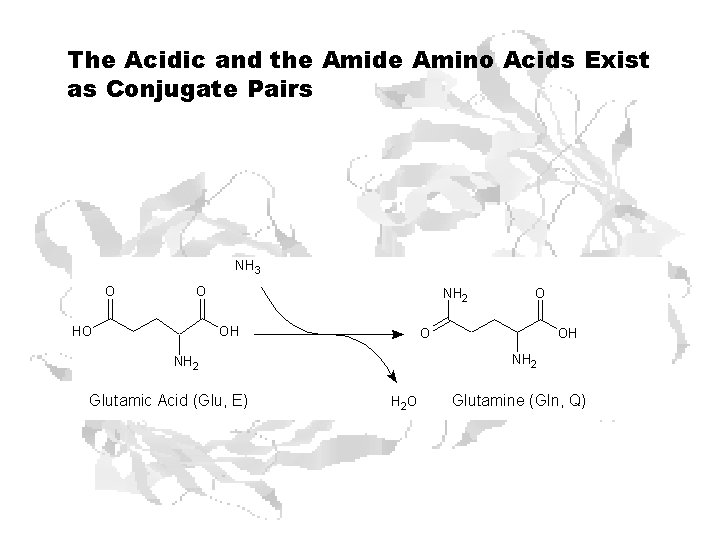
The Acidic and the Amide Amino Acids Exist as Conjugate Pairs




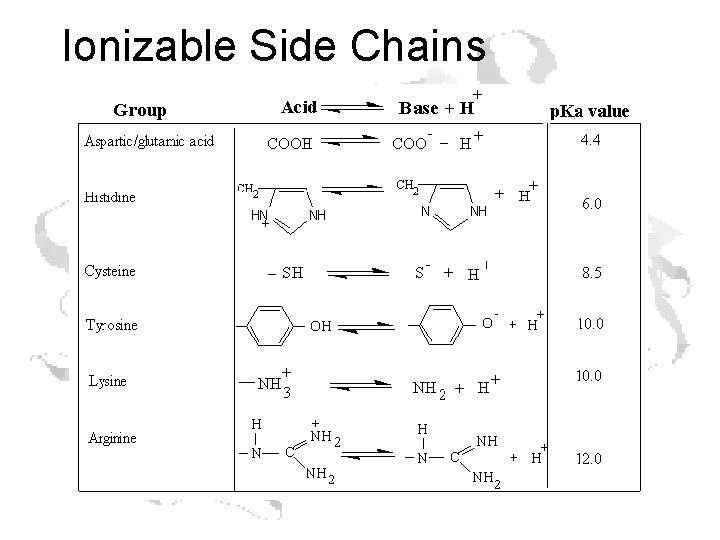
Ionizable Side Chains

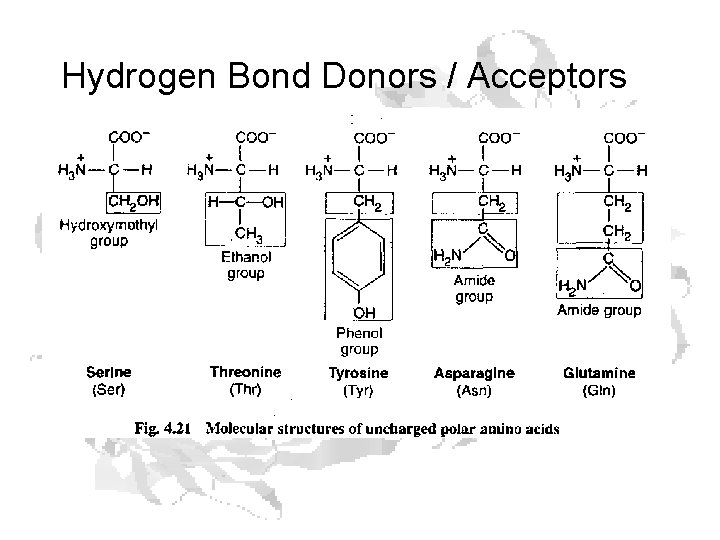
Hydrogen Bond Donors / Acceptors

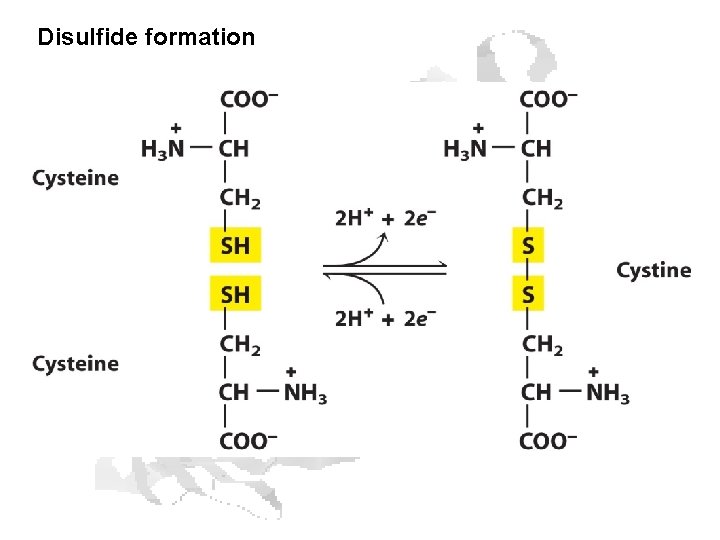
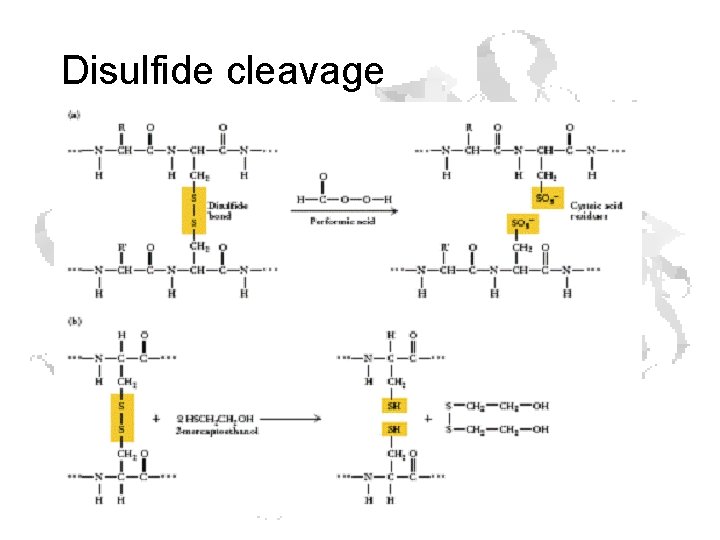
Disulfide formation

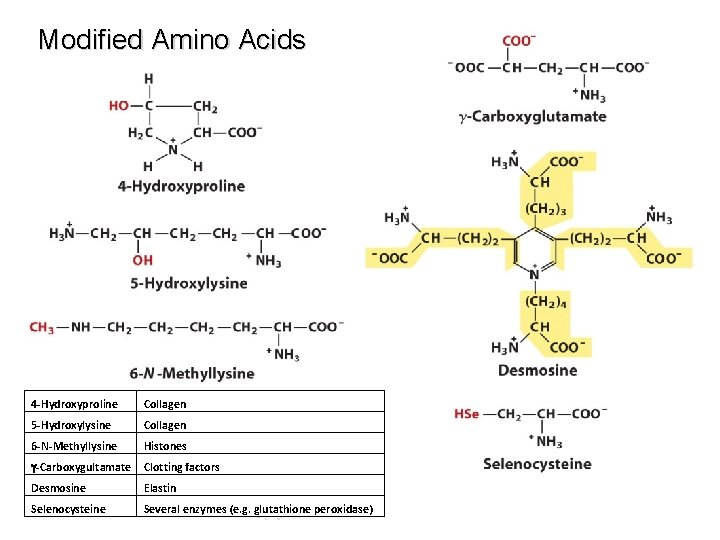
Modified Amino Acids 4 -Hydroxyproline Collagen 5 -Hydroxylysine Collagen 6 -N-Methyllysine Histones g-Carboxygultamate Clotting factors Desmosine Elastin Selenocysteine Several enzymes (e. g. glutathione peroxidase)

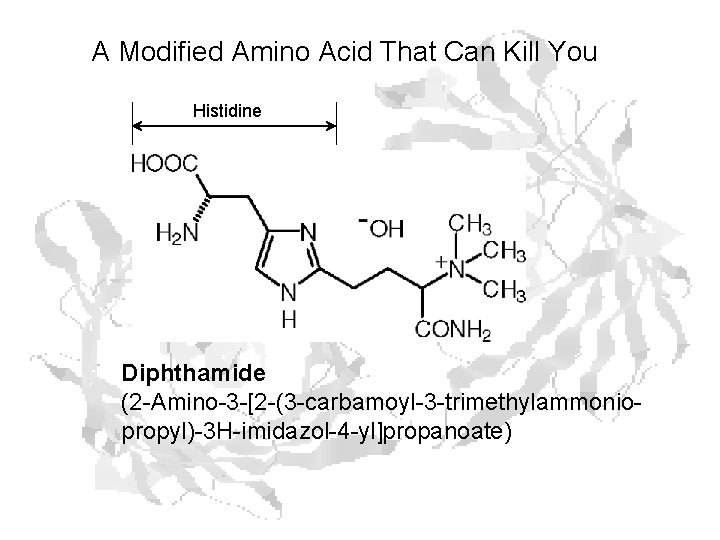
A Modified Amino Acid That Can Kill You Histidine Diphthamide (2 -Amino-3 -[2 -(3 -carbamoyl-3 -trimethylammoniopropyl)-3 H-imidazol-4 -yl]propanoate)

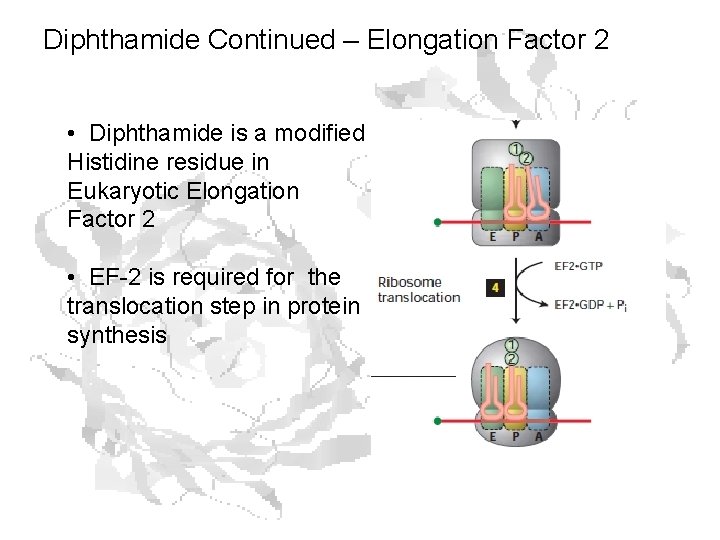
Diphthamide Continued – Elongation Factor 2 • Diphthamide is a modified Histidine residue in Eukaryotic Elongation Factor 2 • EF-2 is required for the translocation step in protein synthesis

Corynebacterium diphtheriae Corynebacteriophage

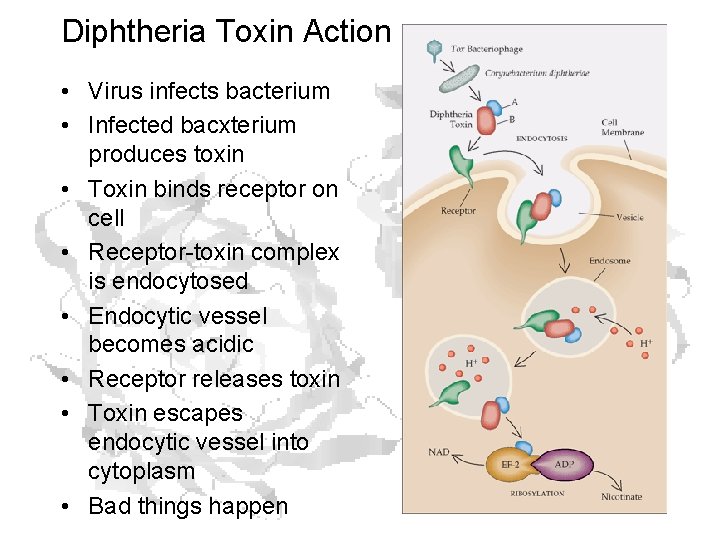
Diphtheria Toxin Action • Virus infects bacterium • Infected bacxterium produces toxin • Toxin binds receptor on cell • Receptor-toxin complex is endocytosed • Endocytic vessel becomes acidic • Receptor releases toxin • Toxin escapes endocytic vessel into cytoplasm • Bad things happen

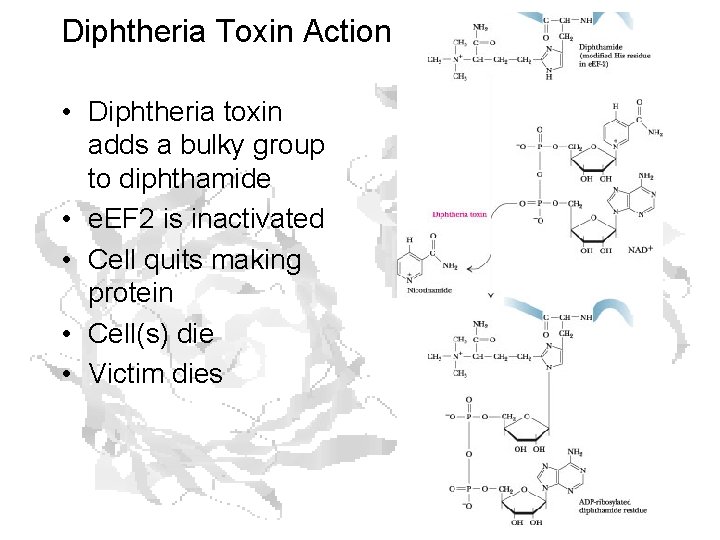
Diphtheria Toxin Action • Diphtheria toxin adds a bulky group to diphthamide • e. EF 2 is inactivated • Cell quits making protein • Cell(s) die • Victim dies

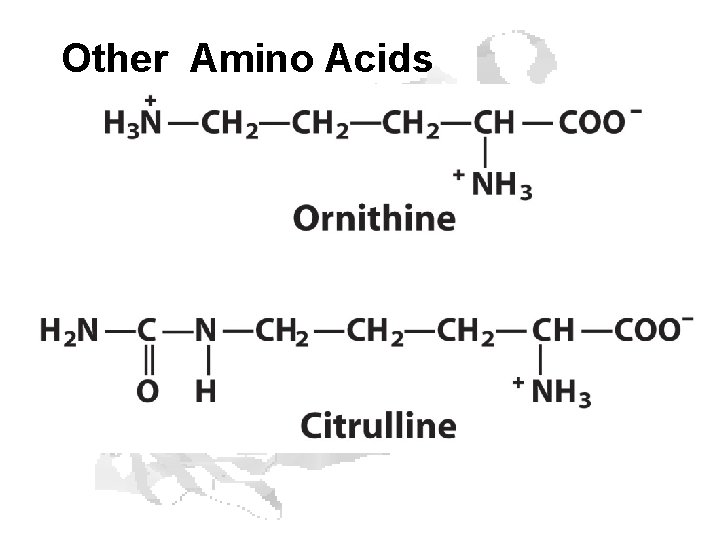
Other Amino Acids


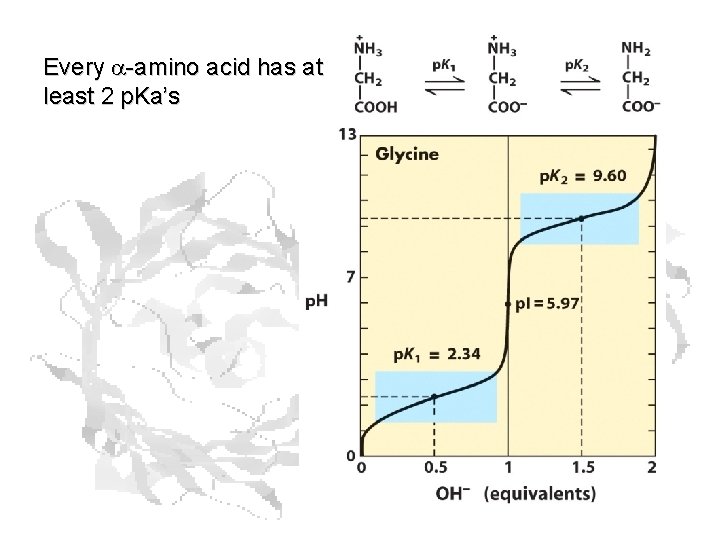
Every a-amino acid has at least 2 p. Ka’s




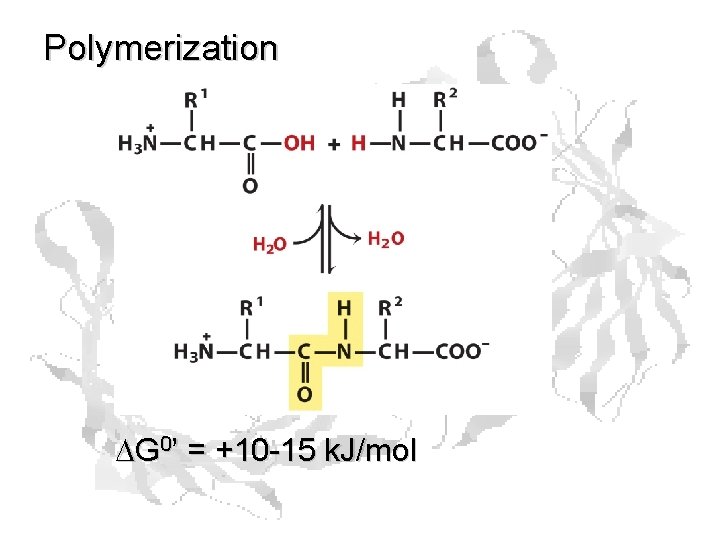
Polymerization DG 0’ = +10 -15 k. J/mol

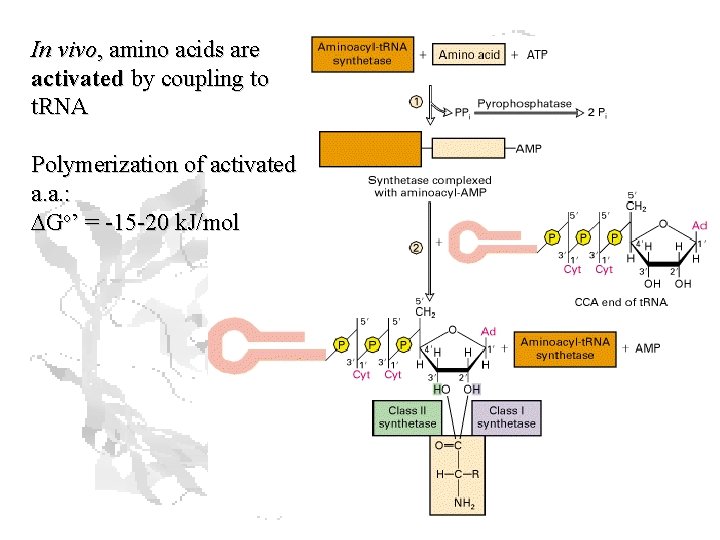
In vivo, amino acids are activated by coupling to t. RNA Polymerization of activated a. a. : DGo’ = -15 -20 k. J/mol

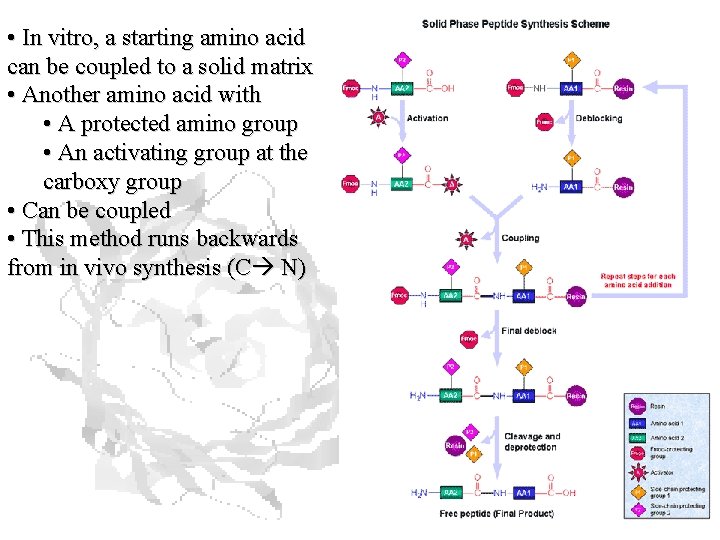
• In vitro, a starting amino acid can be coupled to a solid matrix • Another amino acid with • A protected amino group • An activating group at the carboxy group • Can be coupled • This method runs backwards from in vivo synthesis (C N)

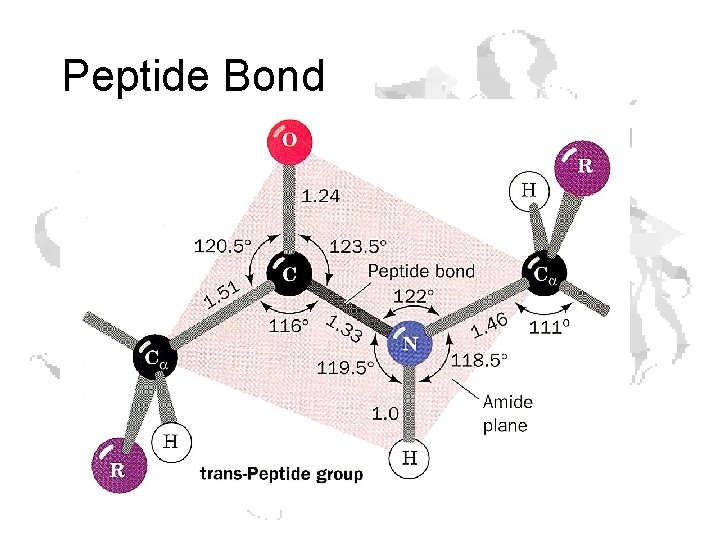
Peptide Bond

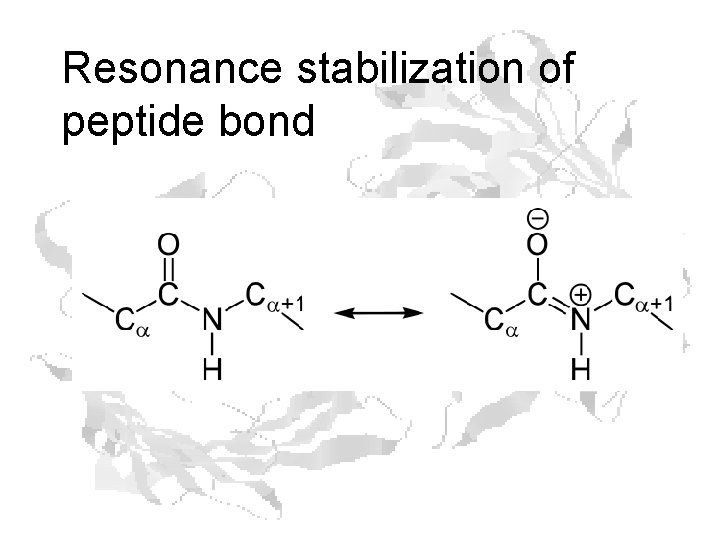
Resonance stabilization of peptide bond

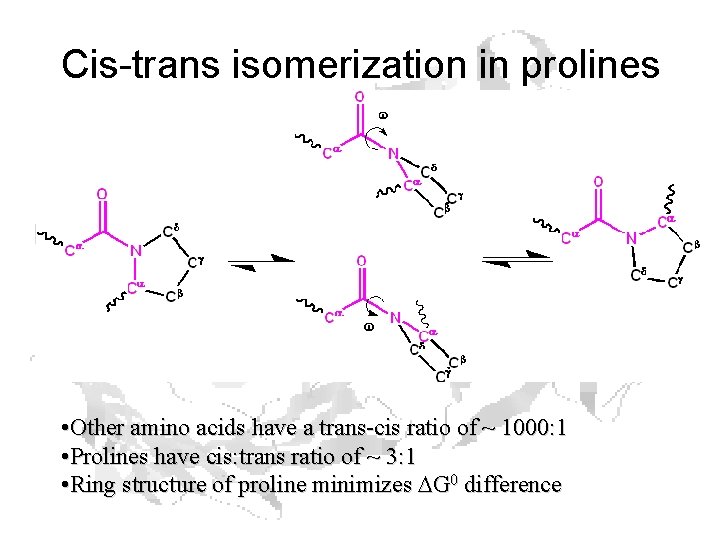
Cis-trans isomerization in prolines • Other amino acids have a trans-cis ratio of ~ 1000: 1 • Prolines have cis: trans ratio of ~ 3: 1 • Ring structure of proline minimizes DG 0 difference







Physical Methods or How to Purify and Sequence a Weapons-Grade Protein

First Question How do I measure the amount of protein I have?

UV Absorption Spectrophotometry


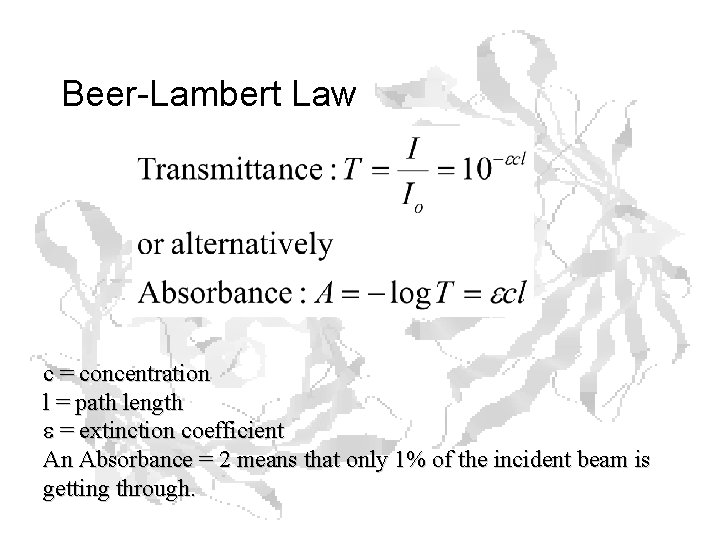
Beer-Lambert Law c = concentration l = path length e = extinction coefficient An Absorbance = 2 means that only 1% of the incident beam is getting through.

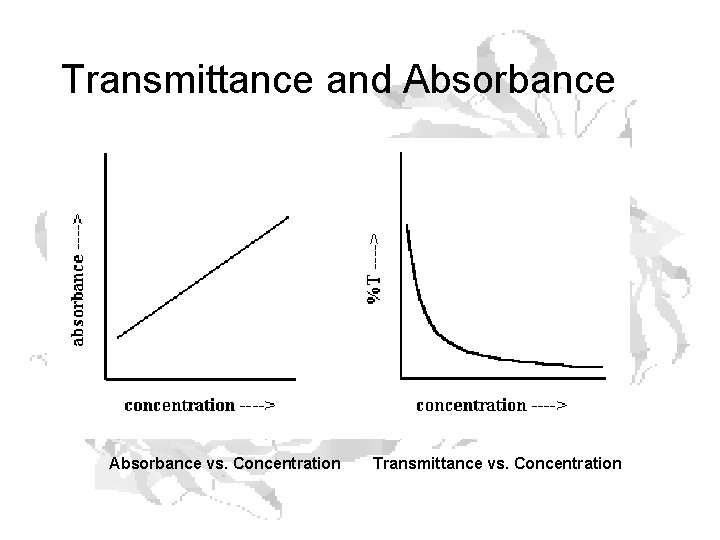
Transmittance and Absorbance vs. Concentration Transmittance vs. Concentration


Second Question How can I spot my protein in the great mass of different proteins?

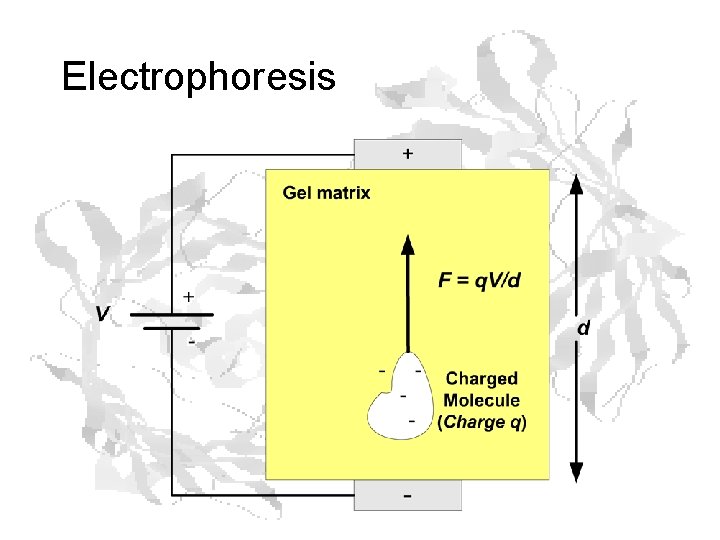
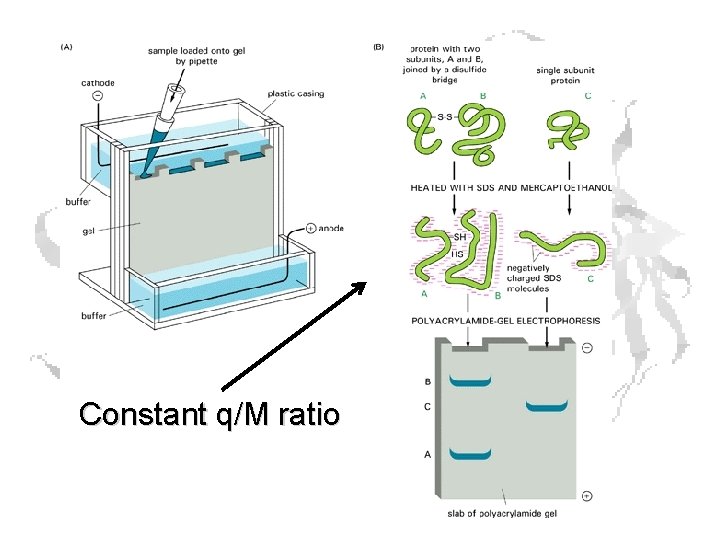
Electrophoresis


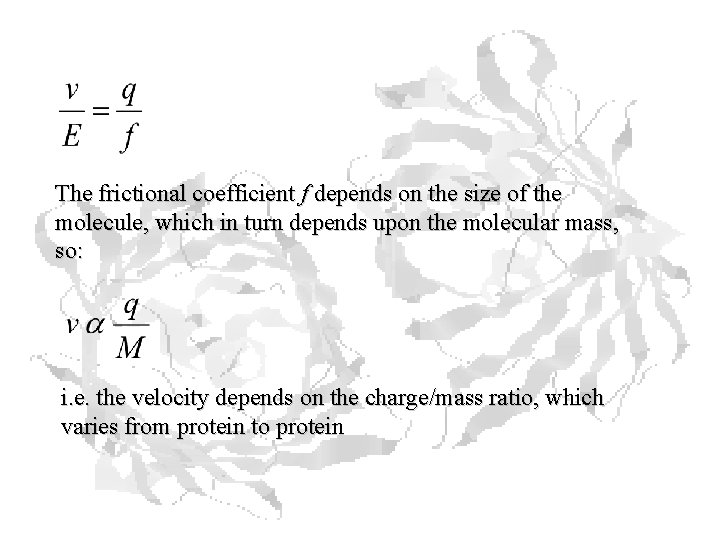
The frictional coefficient f depends on the size of the molecule, which in turn depends upon the molecular mass, so: i. e. the velocity depends on the charge/mass ratio, which varies from protein to protein

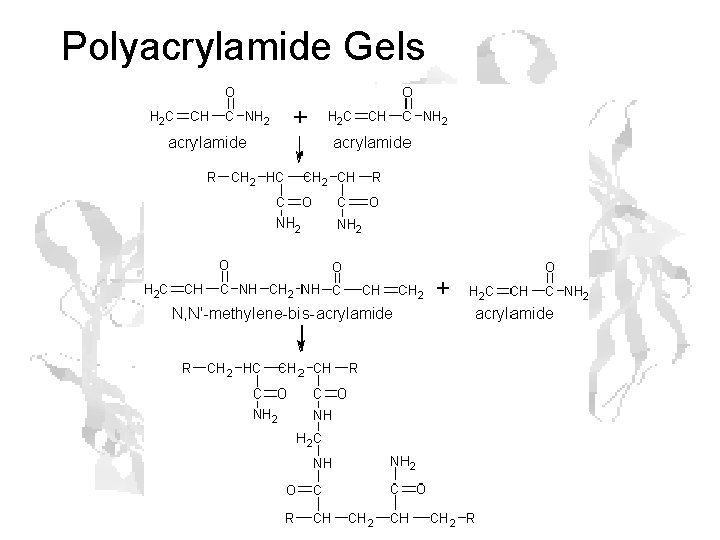
Polyacrylamide Gels


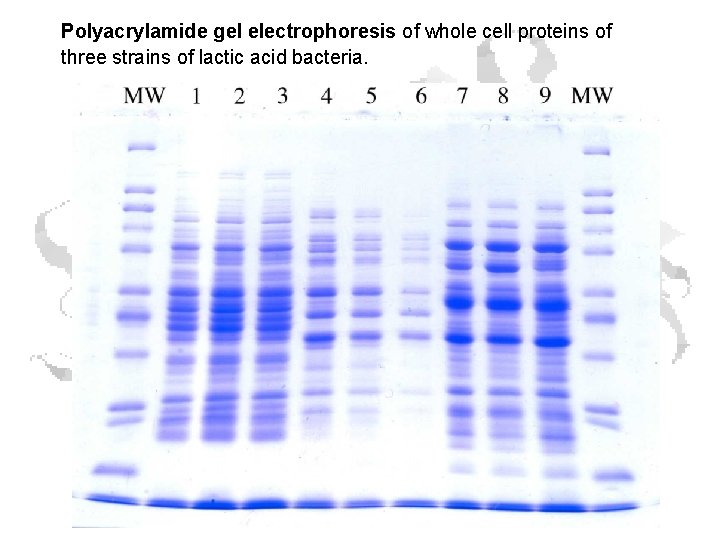
Polyacrylamide gel electrophoresis of whole cell proteins of three strains of lactic acid bacteria.

Agarose Gelidium sp.


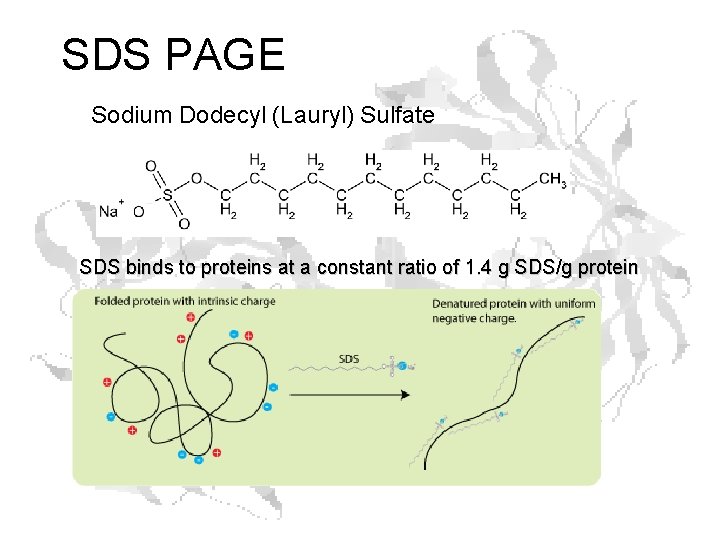
SDS PAGE Sodium Dodecyl (Lauryl) Sulfate SDS binds to proteins at a constant ratio of 1. 4 g SDS/g protein

Constant q/M ratio




Disulfide cleavage

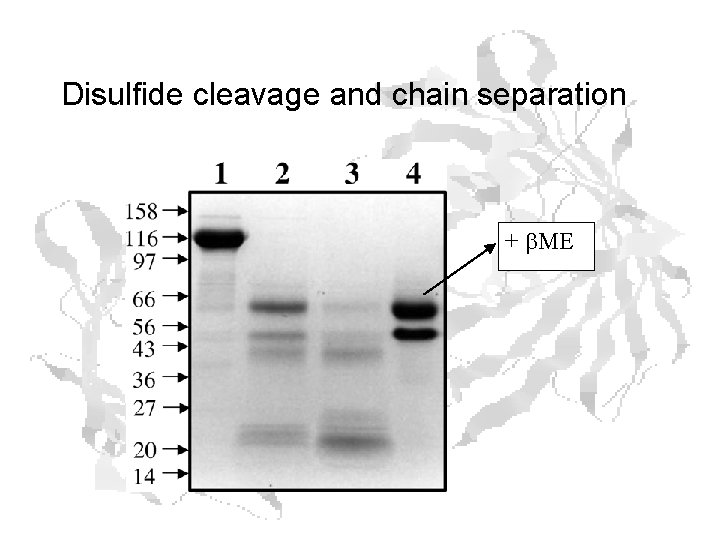
Disulfide cleavage and chain separation + b. ME

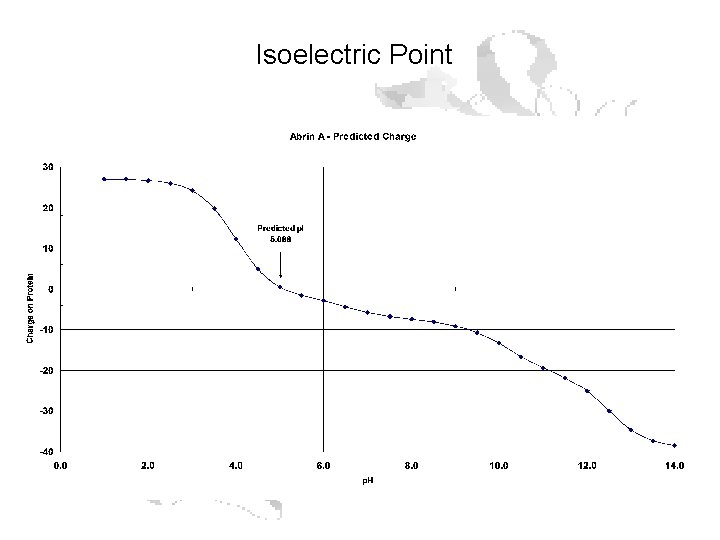
Isoelectric Point


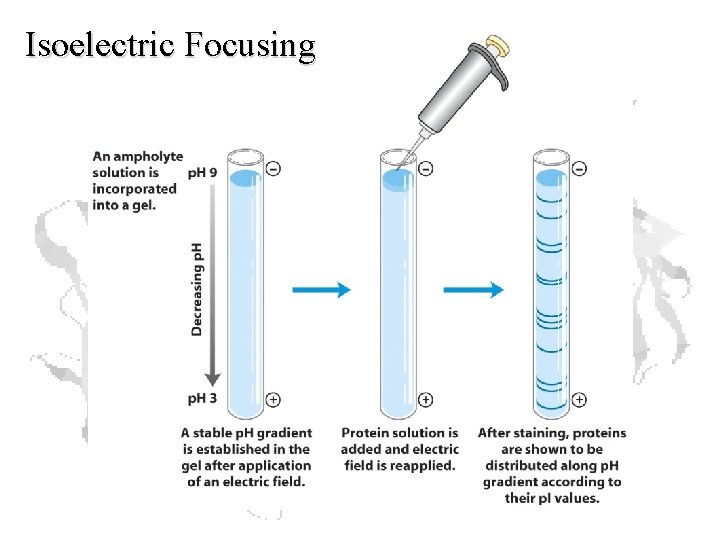
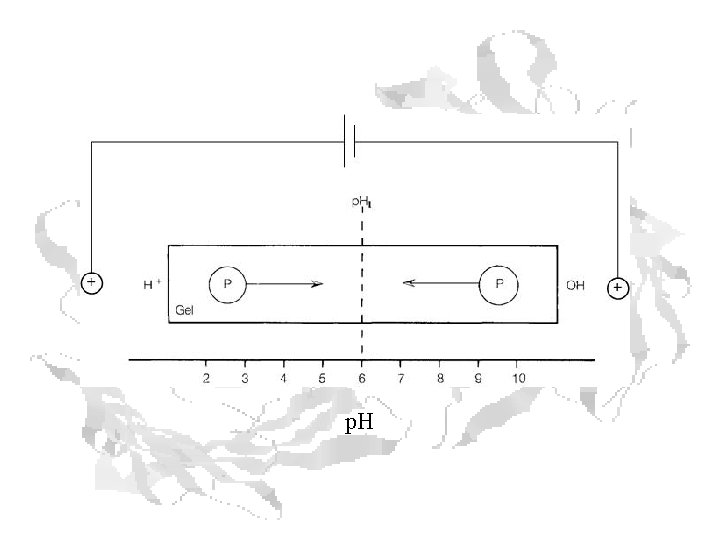
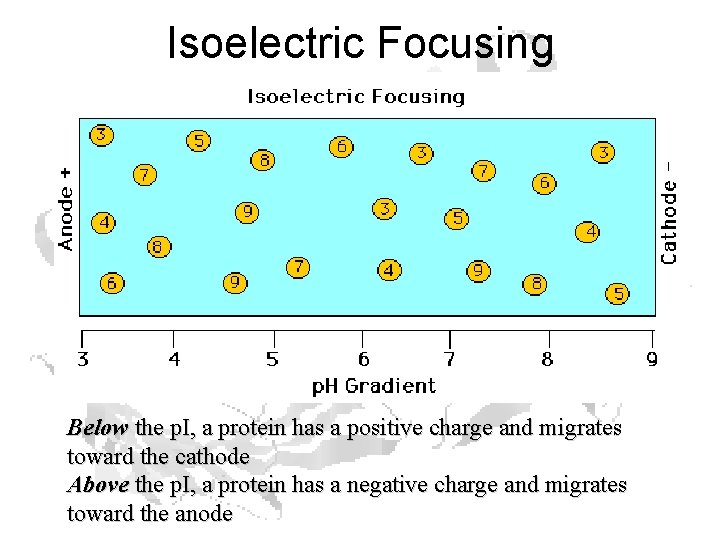
Isoelectric Focusing

p. H

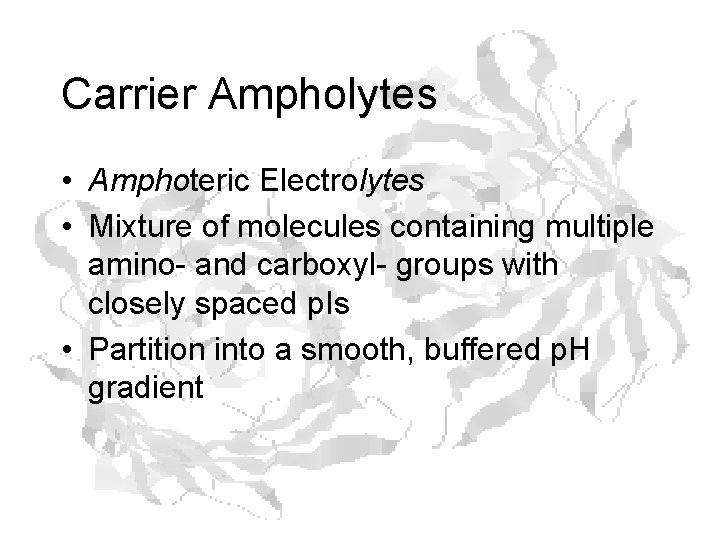
Carrier Ampholytes • Amphoteric Electrolytes • Mixture of molecules containing multiple amino- and carboxyl- groups with closely spaced p. Is • Partition into a smooth, buffered p. H gradient

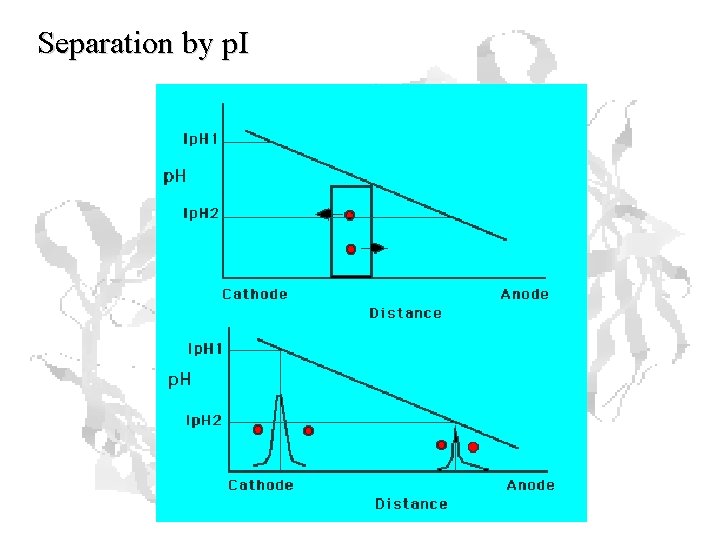
Separation by p. I

Isoelectric Focusing Below the p. I, a protein has a positive charge and migrates toward the cathode Above the p. I, a protein has a negative charge and migrates toward the anode

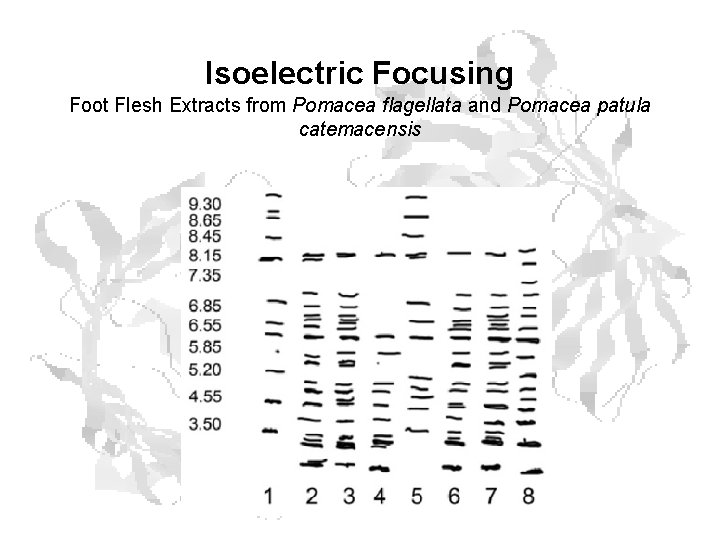
Isoelectric Focusing Foot Flesh Extracts from Pomacea flagellata and Pomacea patula catemacensis

STOP HERE

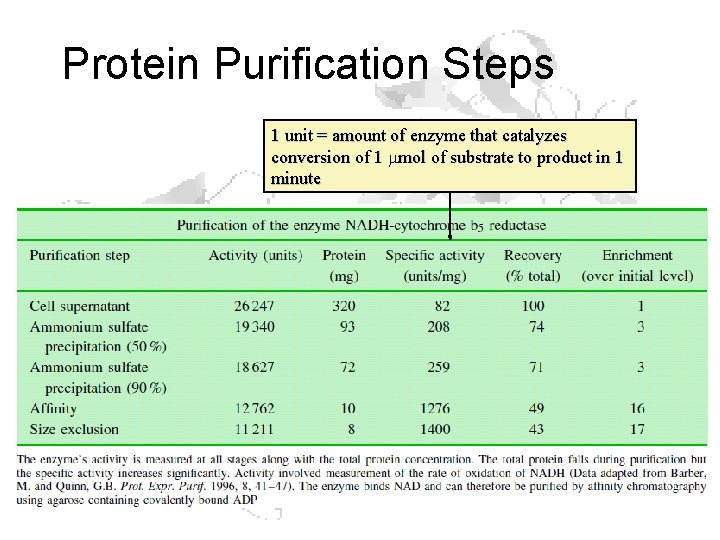
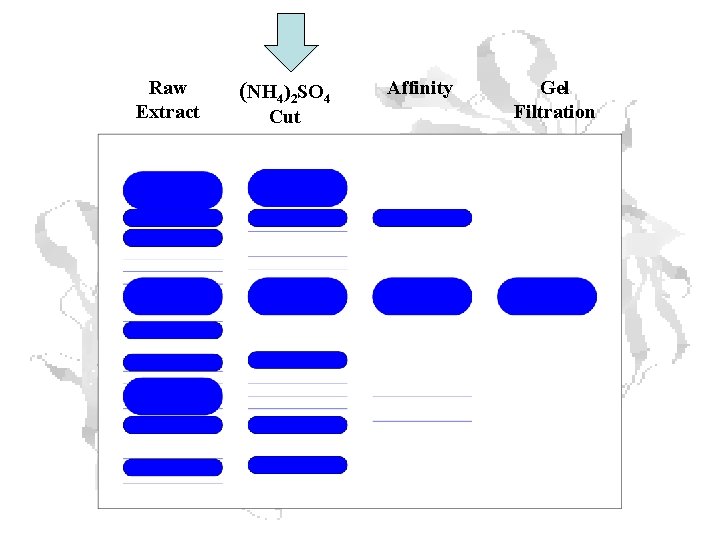
Protein Purification Steps 1 unit = amount of enzyme that catalyzes conversion of 1 mmol of substrate to product in 1 minute

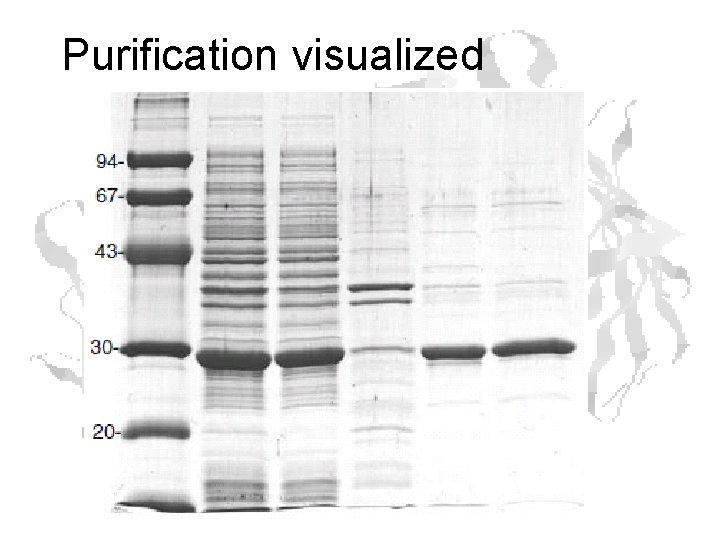
Purification visualized

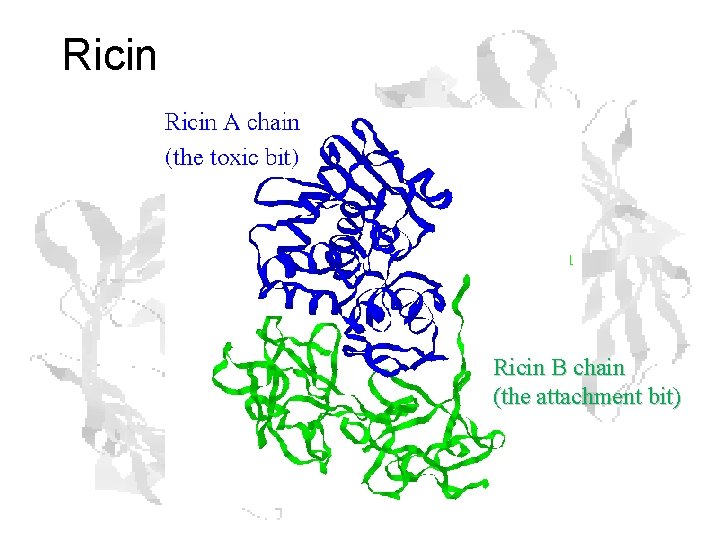
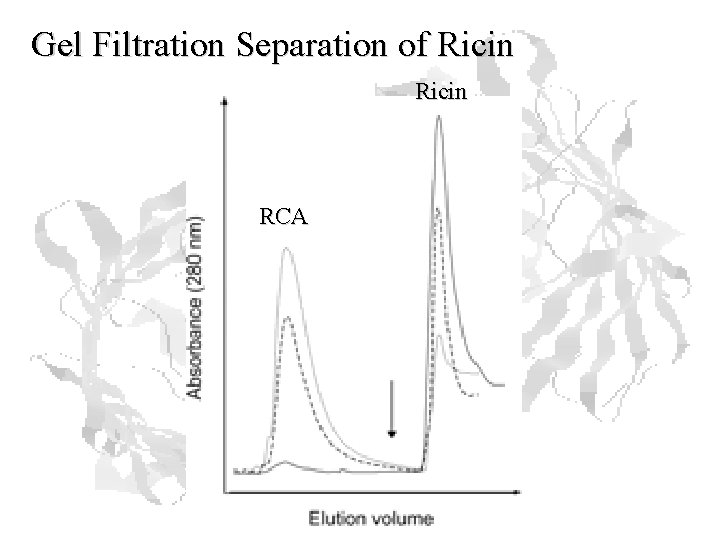
Example: Purification of Ricin

Georgi Markov 1929 -1978


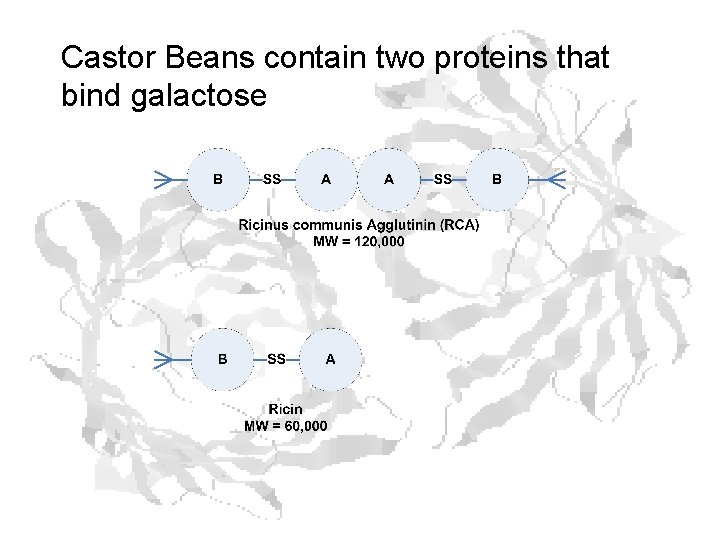
Ricinus communis – castor oil plant

Ricin B chain (the attachment bit)

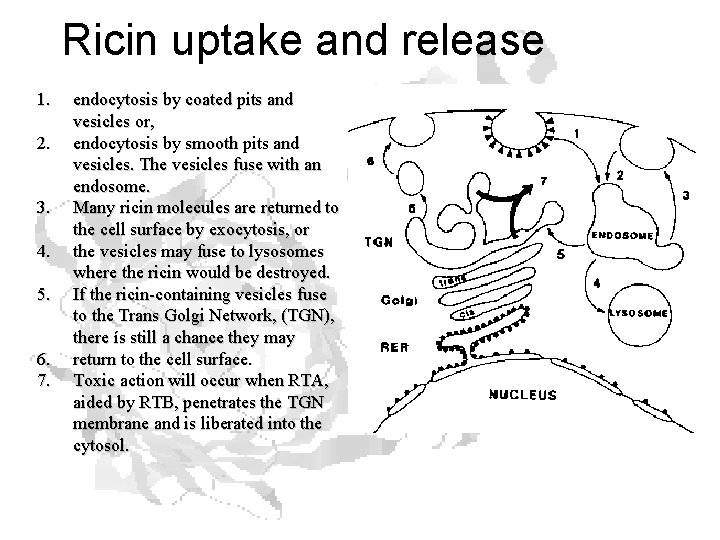
Ricin uptake and release 1. 2. 3. 4. 5. 6. 7. endocytosis by coated pits and vesicles or, endocytosis by smooth pits and vesicles. The vesicles fuse with an endosome. Many ricin molecules are returned to the cell surface by exocytosis, or the vesicles may fuse to lysosomes where the ricin would be destroyed. If the ricin-containing vesicles fuse to the Trans Golgi Network, (TGN), there ís still a chance they may return to the cell surface. Toxic action will occur when RTA, aided by RTB, penetrates the TGN membrane and is liberated into the cytosol.

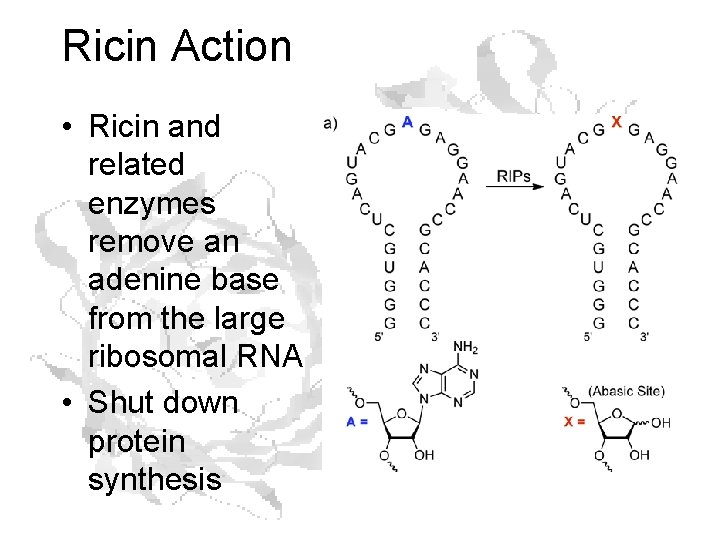
Ricin Action • Ricin and related enzymes remove an adenine base from the large ribosomal RNA • Shut down protein synthesis

The possibility that ricin might be used as an asymmetric warfare weapon has not escaped the attention of the armed services. The last time I was qualified to know for sure, there were no effective antidotes.

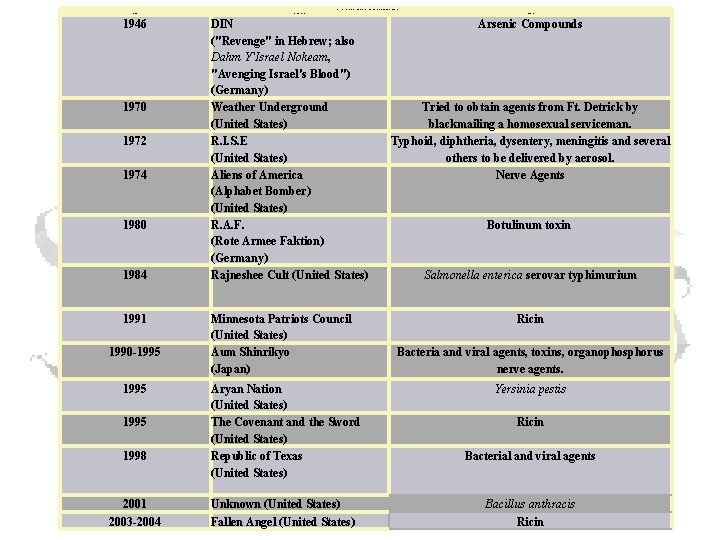
Significant Terrorist Incidents Involving Chemical and Biological Agents Year 1946 1970 1972 1974 1980 1984 1991 1990 -1995 1998 2001 2003 -2004 Organization DIN ("Revenge" in Hebrew; also Dahm Y'Israel Nokeam, "Avenging Israel's Blood") (Germany) Weather Underground (United States) R. I. S. E (United States) Aliens of America (Alphabet Bomber) (United States) R. A. F. (Rote Armee Faktion) (Germany) Rajneshee Cult (United States) Minnesota Patriots Council (United States) Aum Shinrikyo (Japan) Aryan Nation (United States) The Covenant and the Sword (United States) Republic of Texas (United States) Unknown (United States) Fallen Angel (United States) Agents Arsenic Compounds Tried to obtain agents from Ft. Detrick by blackmailing a homosexual serviceman. Typhoid, diphtheria, dysentery, meningitis and several others to be delivered by aerosol. Nerve Agents Botulinum toxin Salmonella enterica serovar typhimurium Ricin Bacteria and viral agents, toxins, organophosphorus nerve agents. Yersinia pestis Ricin Bacterial and viral agents Bacillus anthracis Ricin

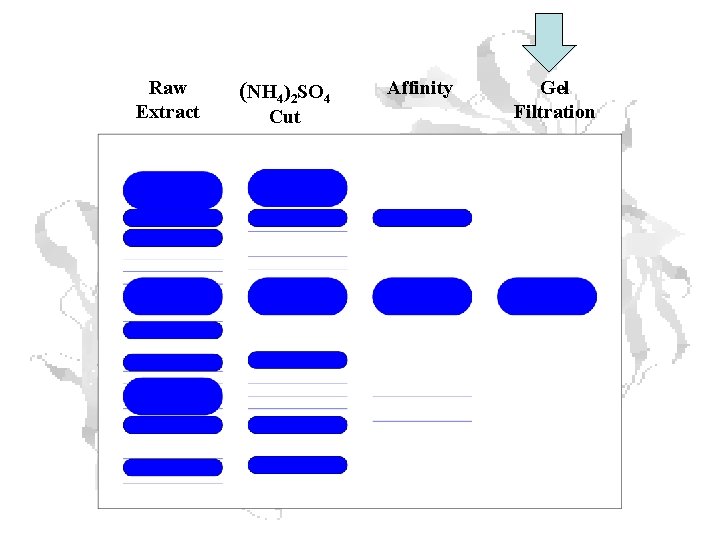
Raw Extract (NH 4)2 SO 4 Cut Affinity Gel Filtration

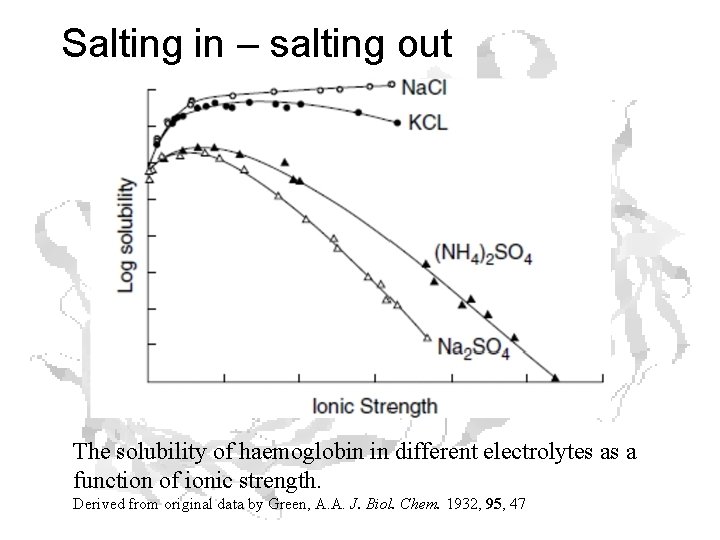
Salting In – Salting out • salting in: Increasing ionic strength increases protein solubility • salting out: Increasing further leads to a loss of solubility

Salting in – salting out The solubility of haemoglobin in different electrolytes as a function of ionic strength. Derived from original data by Green, A. A. J. Biol. Chem. 1932, 95, 47

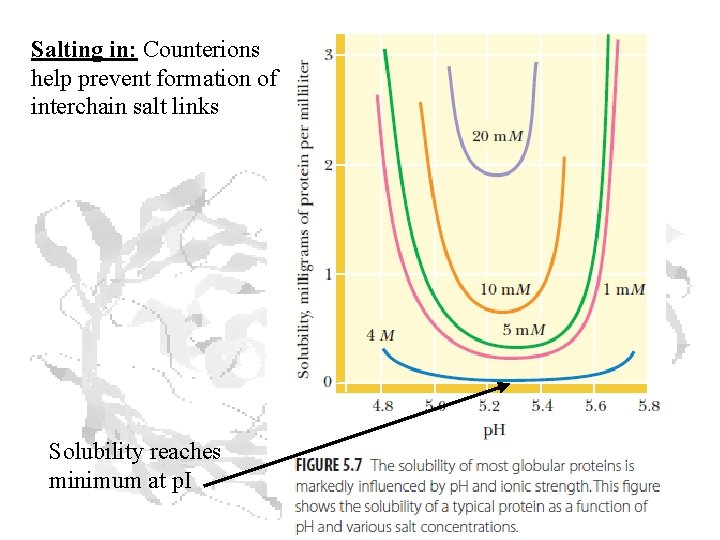
Salting in: Counterions help prevent formation of interchain salt links Solubility reaches minimum at p. I

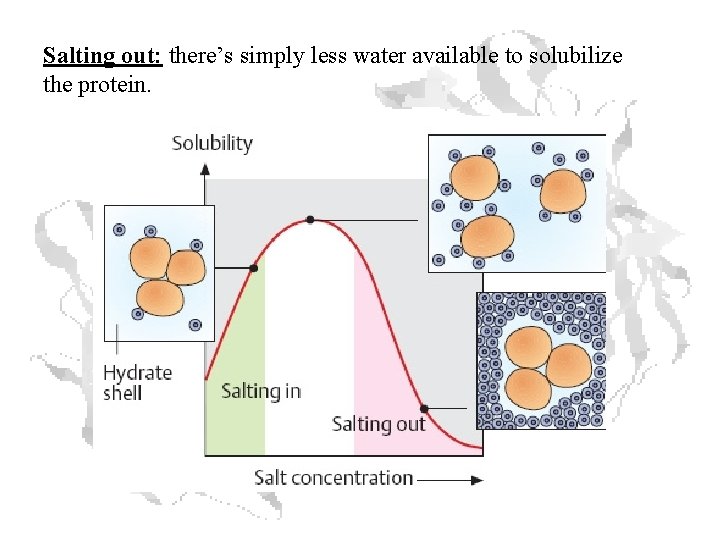
Salting out: there’s simply less water available to solubilize the protein.

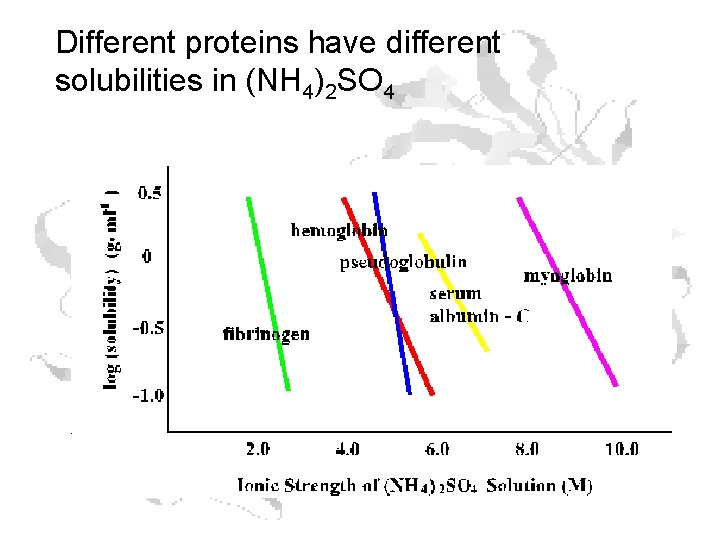
Different proteins have different solubilities in (NH 4)2 SO 4

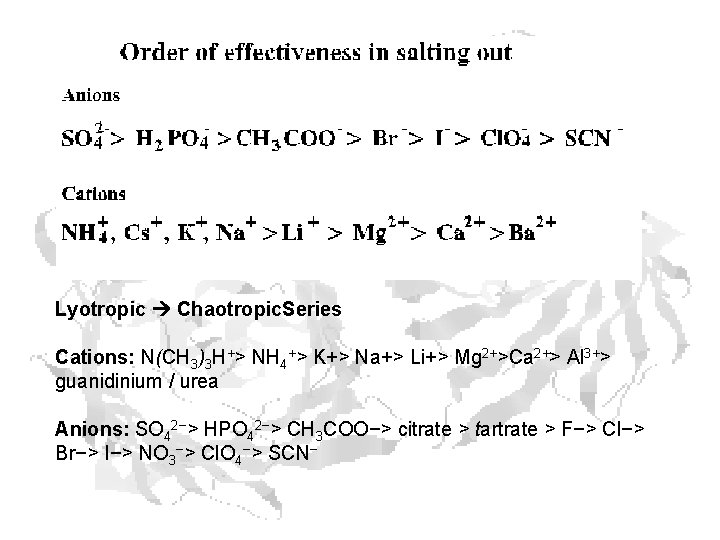
Lyotropic Chaotropic. Series Cations: N(CH 3)3 H+> NH 4+> K+> Na+> Li+> Mg 2+>Ca 2+> Al 3+> guanidinium / urea Anions: SO 42−> HPO 42−> CH 3 COO−> citrate > tartrate > F−> Cl−> Br−> I−> NO 3−> Cl. O 4−> SCN−

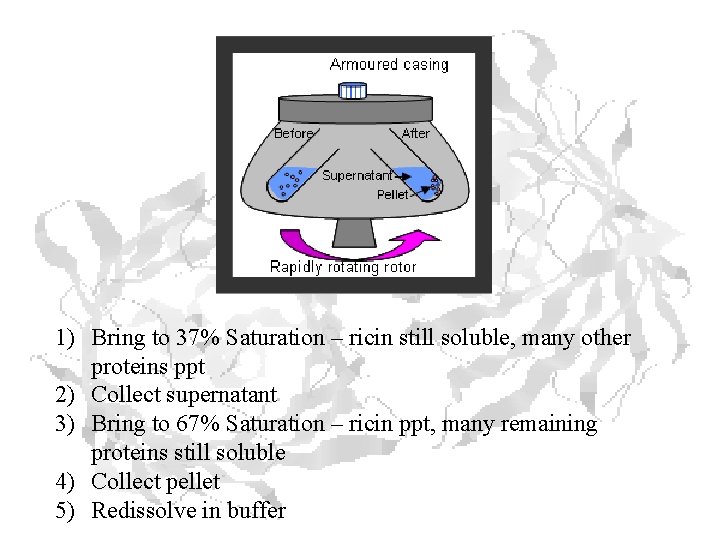
1) Bring to 37% Saturation – ricin still soluble, many other proteins ppt 2) Collect supernatant 3) Bring to 67% Saturation – ricin ppt, many remaining proteins still soluble 4) Collect pellet 5) Redissolve in buffer

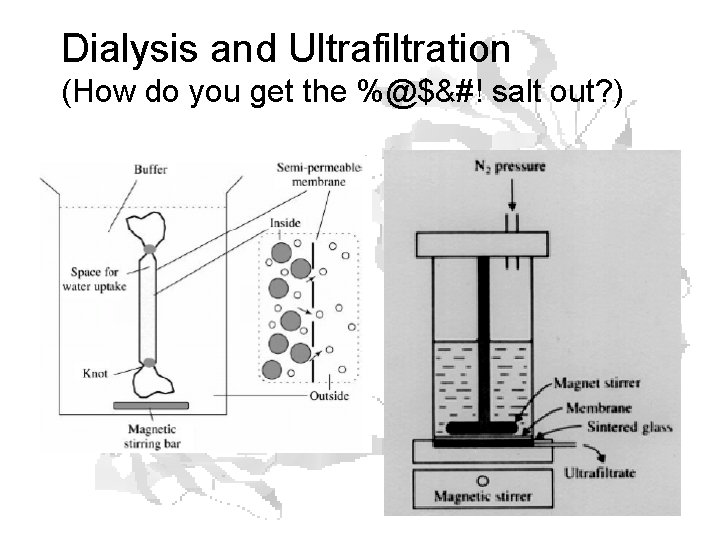
Dialysis and Ultrafiltration (How do you get the %@$&#! salt out? )

Raw Extract (NH 4)2 SO 4 Cut Affinity Gel Filtration

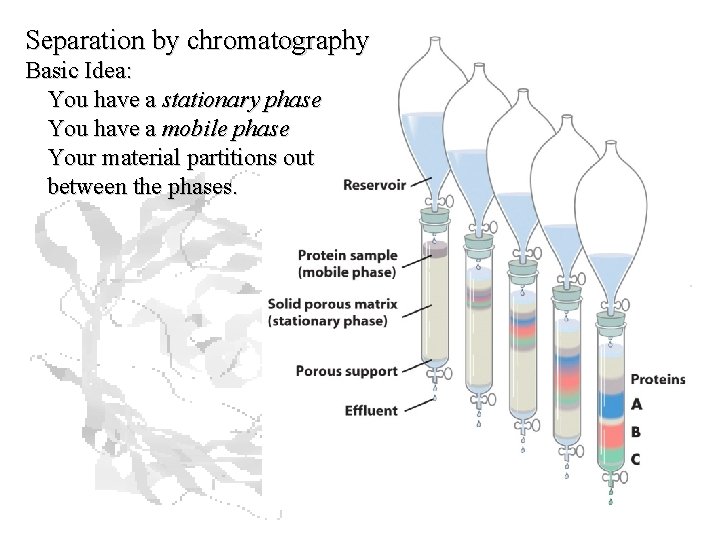
Separation by chromatography Basic Idea: You have a stationary phase You have a mobile phase Your material partitions out between the phases.

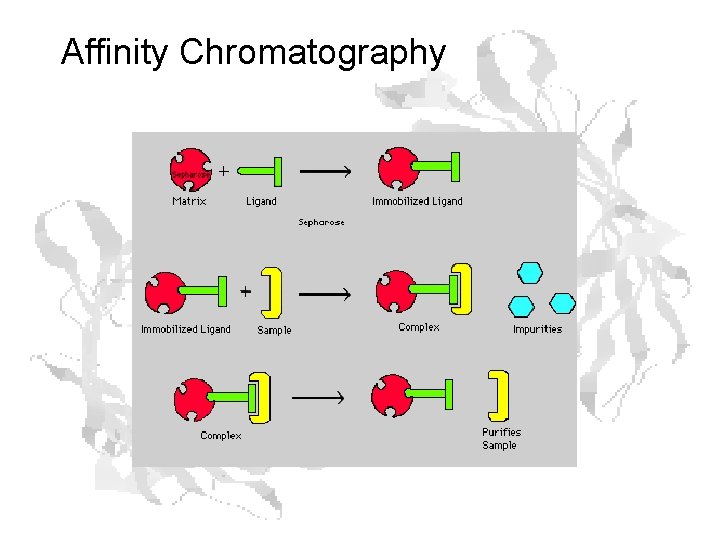
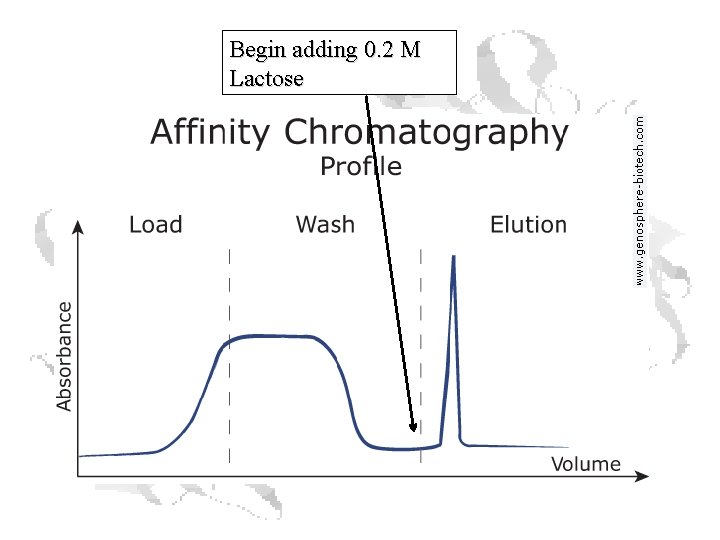
Affinity Chromatography


Structure of Agarose is a polymer of agarobiose, which in turn consists of one unit each of galactose and 3, 6 -anhydro-a-L-galactose. Ricin sticks to galactose, so store-bought agarose acts as an affinity column right out of the bottle, with ricin binding the beads while other proteins wash through.

Begin adding 0. 2 M Lactose

Raw Extract (NH 4)2 SO 4 Cut Affinity Gel Filtration

Castor Beans contain two proteins that bind galactose

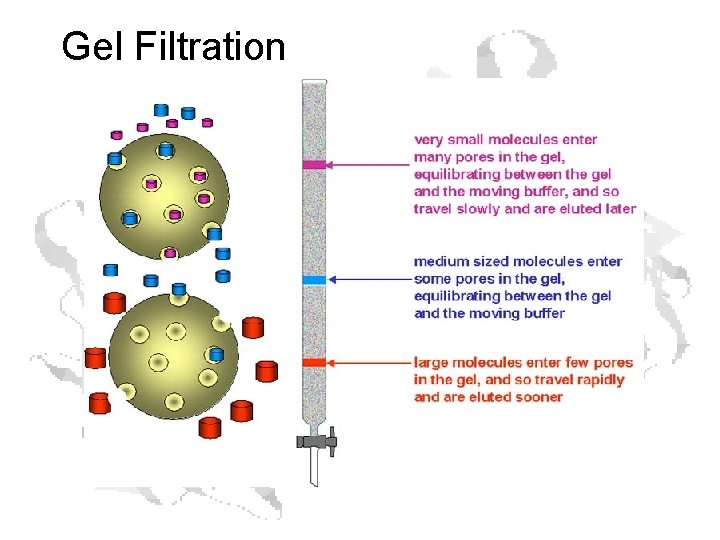
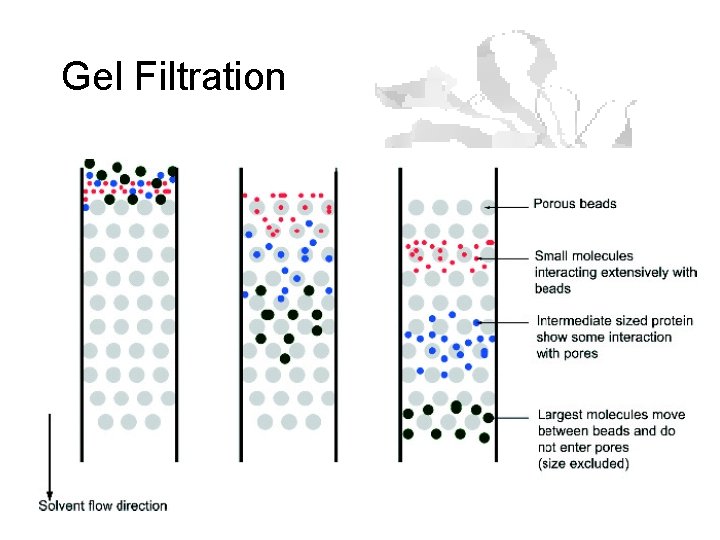
Gel Filtration

Gel Filtration

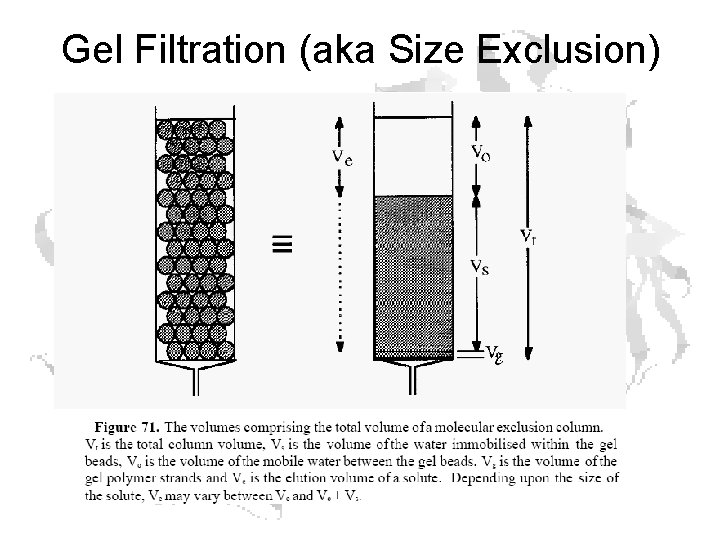
Gel Filtration (aka Size Exclusion)

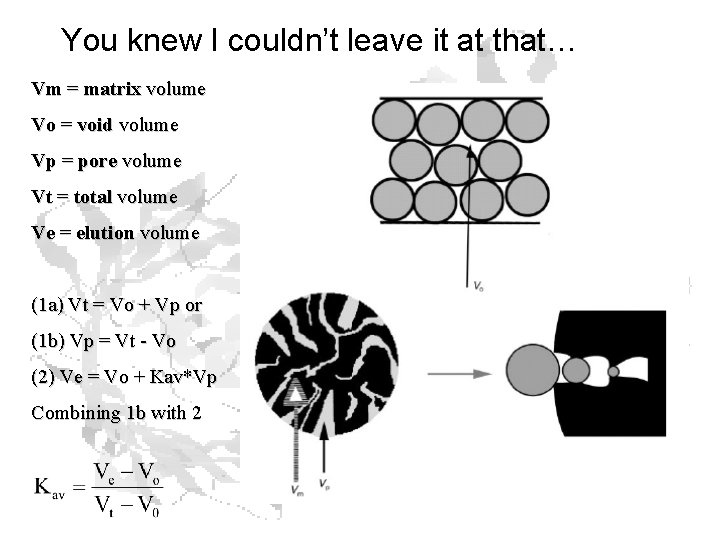
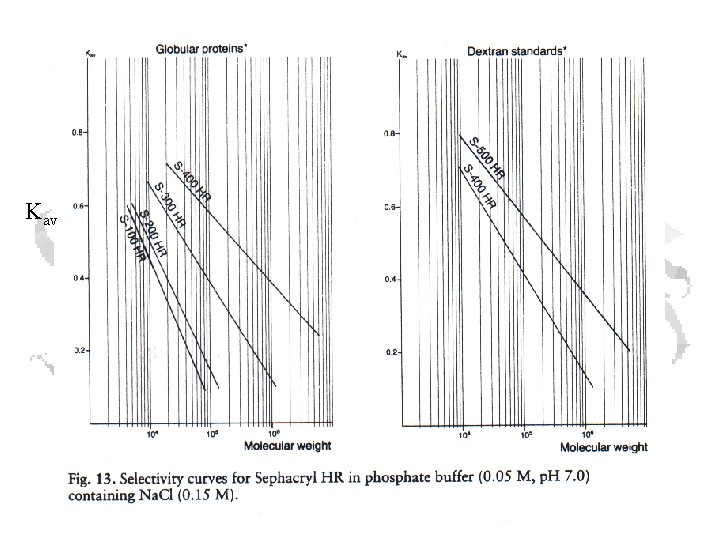
You knew I couldn’t leave it at that… Vm = matrix volume Vo = void volume Vp = pore volume Vt = total volume Ve = elution volume (1 a) Vt = Vo + Vp or (1 b) Vp = Vt - Vo (2) Ve = Vo + Kav*Vp Combining 1 b with 2

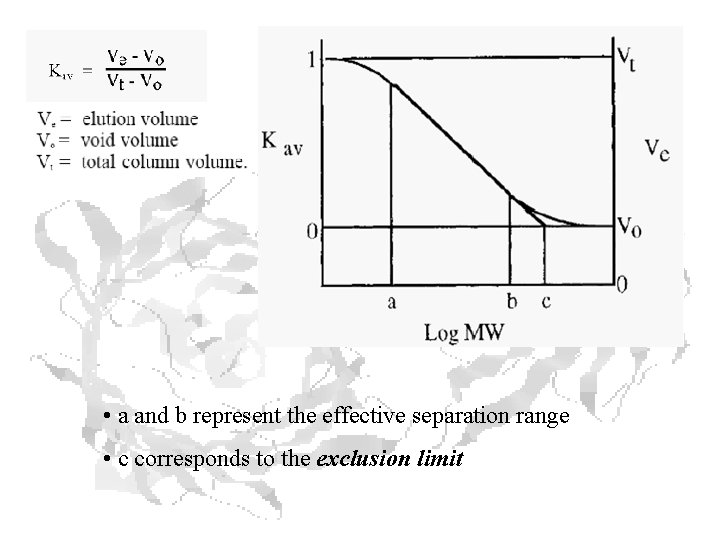
• a and b represent the effective separation range • c corresponds to the exclusion limit

Kav

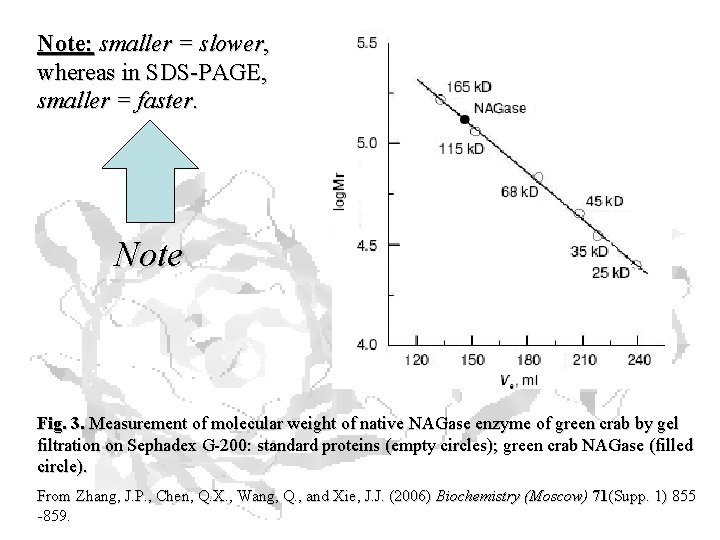
Note: smaller = slower, whereas in SDS-PAGE, smaller = faster. Note Fig. 3. Measurement of molecular weight of native NAGase enzyme of green crab by gel filtration on Sephadex G-200: standard proteins (empty circles); green crab NAGase (filled circle). From Zhang, J. P. , Chen, Q. X. , Wang, Q. , and Xie, J. J. (2006) Biochemistry (Moscow) 71(Supp. 1) 855 -859.

Gel Filtration Separation of Ricin RCA

Raw Extract (NH 4)2 SO 4 Cut Affinity Gel Filtration

Okay, Now Let’s Sequence the A-Chain

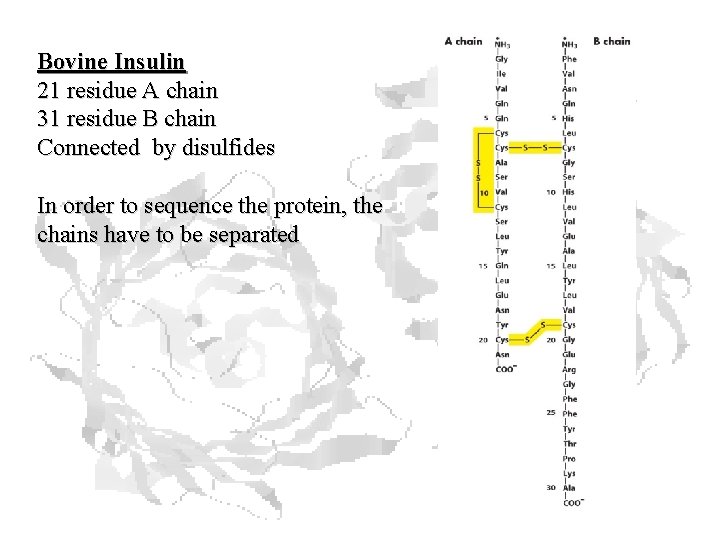
Bovine Insulin 21 residue A chain 31 residue B chain Connected by disulfides In order to sequence the protein, the chains have to be separated


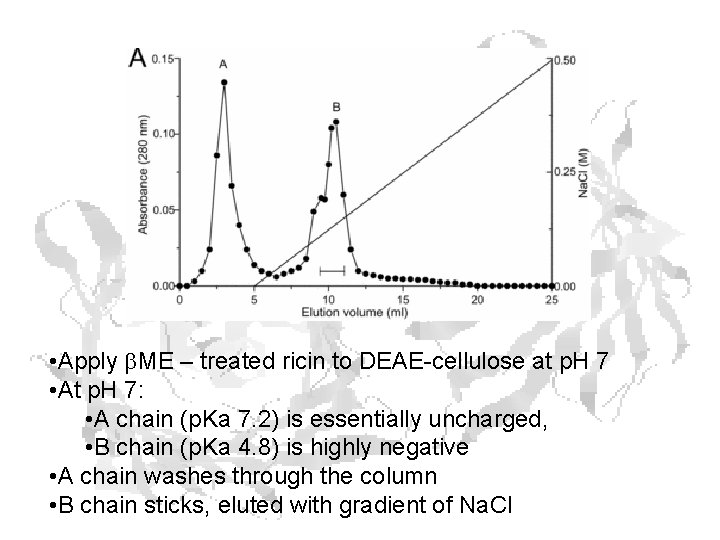
Chain Separation • Interchain disulfide broken by high concentrations of b. ME • Chains are about the same size – but can take advantage of different p. Is – B-Chain – A-Chain p. I ~ 5. 3 p. I ~ 7. 2

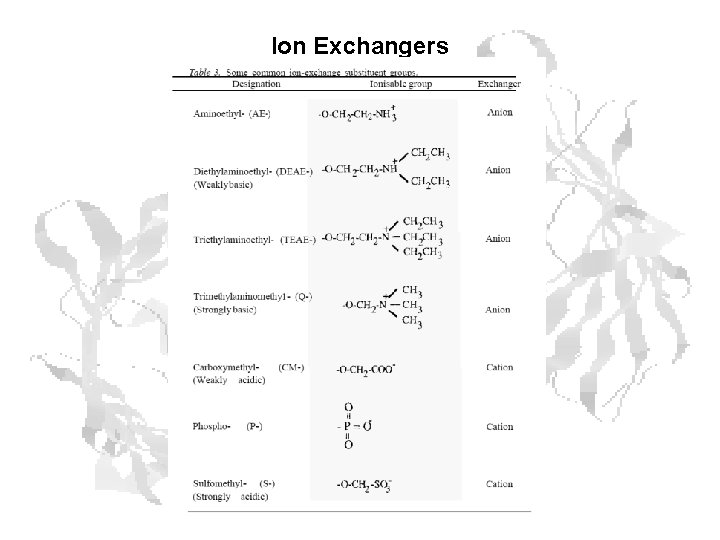
Ion Exchangers


• Apply b. ME – treated ricin to DEAE-cellulose at p. H 7 • At p. H 7: • A chain (p. Ka 7. 2) is essentially uncharged, • B chain (p. Ka 4. 8) is highly negative • A chain washes through the column • B chain sticks, eluted with gradient of Na. Cl

2 -D Electrophoresis (an aside) • Can use two different properties of a protein to separate electrophoretically • For analysis of cellular protein content, often use 2 -dimensional electrophoresis: • 1 st dimension is isoelectric focusing • 2 nd dimension is SDS PAGE




2 -D Electrophoresis (cont. ) • Can use other protein properties to separate – Simple PAGE at 2 different p. Hs – PAGE and SDS PAGE

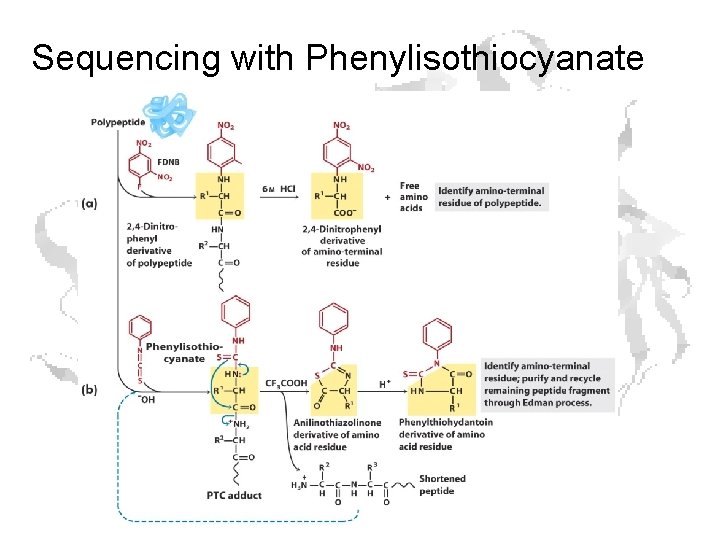
Sequencing with Phenylisothiocyanate

• Applied Biosystems 492 Procise Protein Sequencer


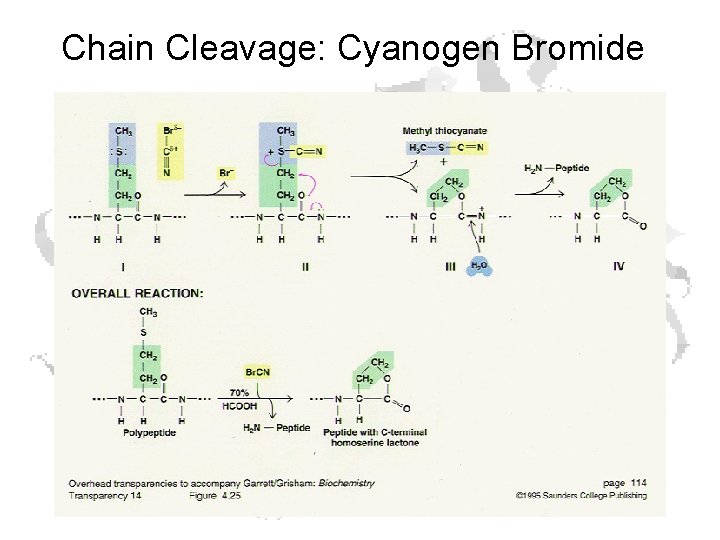
Chain Cleavage: Cyanogen Bromide


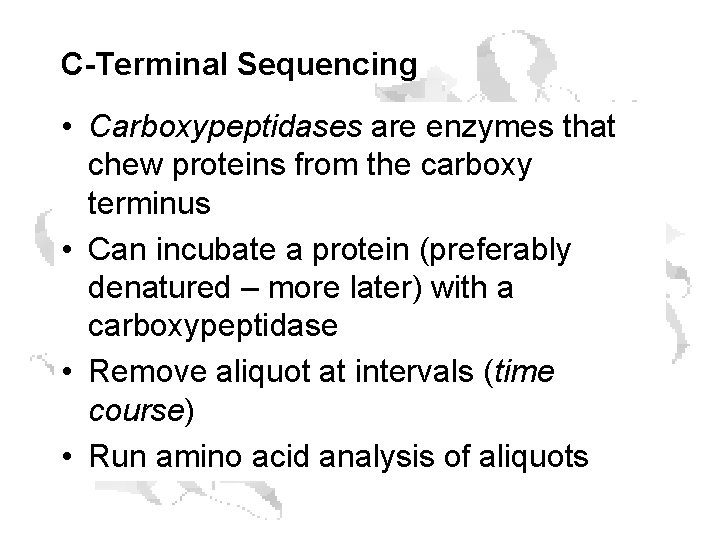
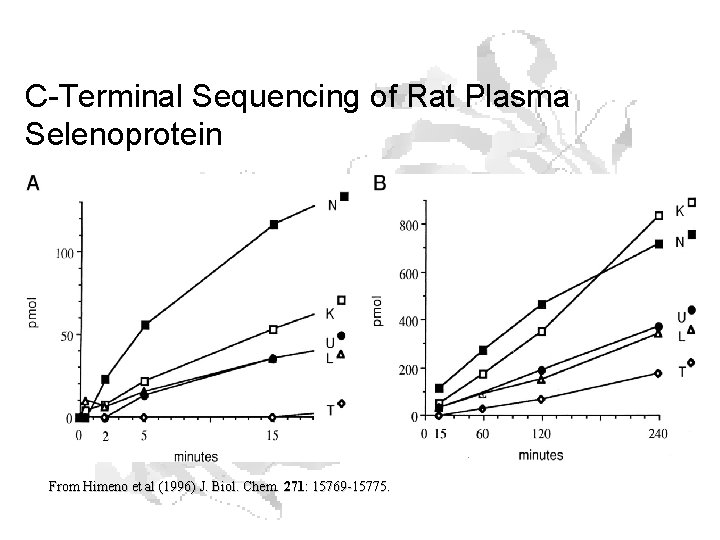
C-Terminal Sequencing • Carboxypeptidases are enzymes that chew proteins from the carboxy terminus • Can incubate a protein (preferably denatured – more later) with a carboxypeptidase • Remove aliquot at intervals (time course) • Run amino acid analysis of aliquots

C-Terminal Sequencing of Rat Plasma Selenoprotein From Himeno et al (1996) J. Biol. Chem. 271: 15769 -15775.

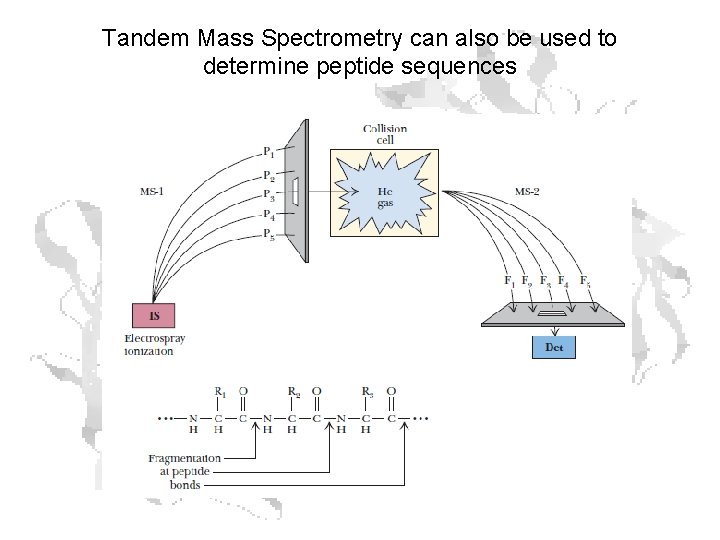
Tandem Mass Spectrometry can also be used to determine peptide sequences

MOLECULAR EVOLUTION


Neutral Theory of Molecular Evolution • Kimura (1968) • Mutations can be: – Advantageous – Detrimental – Neutral (no good or bad phenotypic effect) • Advantageous mutations are rapidly fixed, but really rare • Diadvantageous mutations are rapidly eliminated • Neutral mutations accumulate

What Happens to a Neutral Mutation? • Frequency subject to random chance • Will carrier of gene reproduce? • Many born but few survive – Partly selection – Mostly dumb luck • Gene can have two fates – Elimination (frequent – Fixation (rare)

Genetic Drift in Action Our green genes are evolutionarily superior! Ow! Never mind…

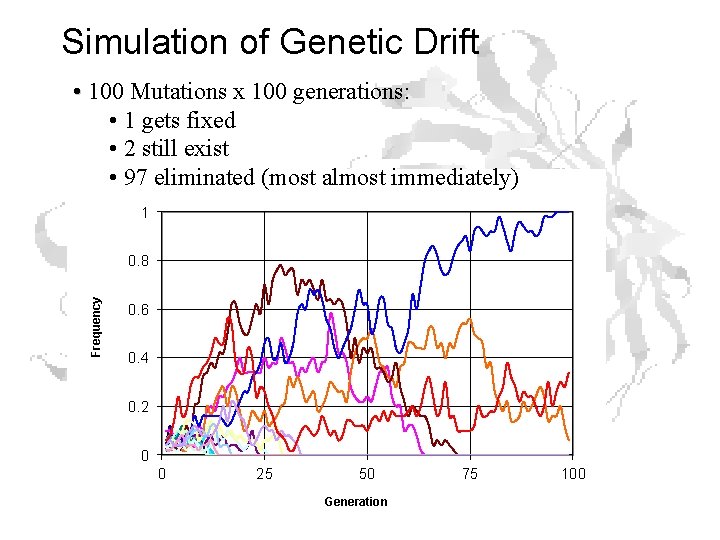
Simulation of Genetic Drift • 100 Mutations x 100 generations: • 1 gets fixed • 2 still exist • 97 eliminated (most almost immediately) 1 Frequency 0. 8 0. 6 0. 4 0. 2 0 0 25 50 Generation 75 100

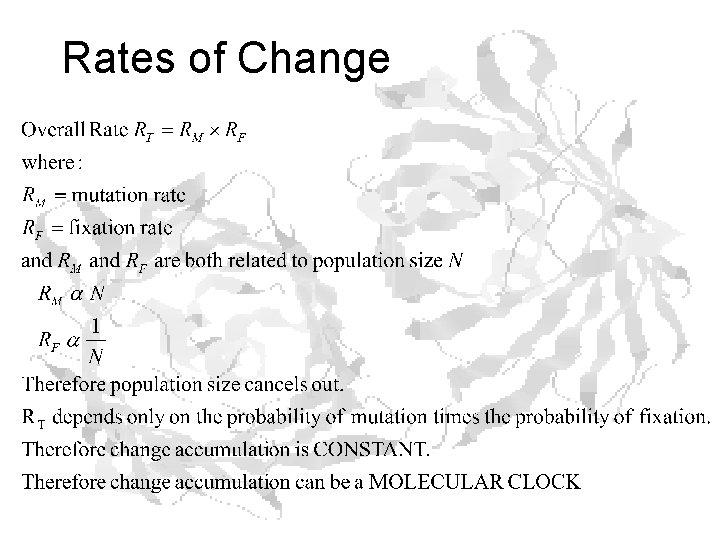
Rates of Change

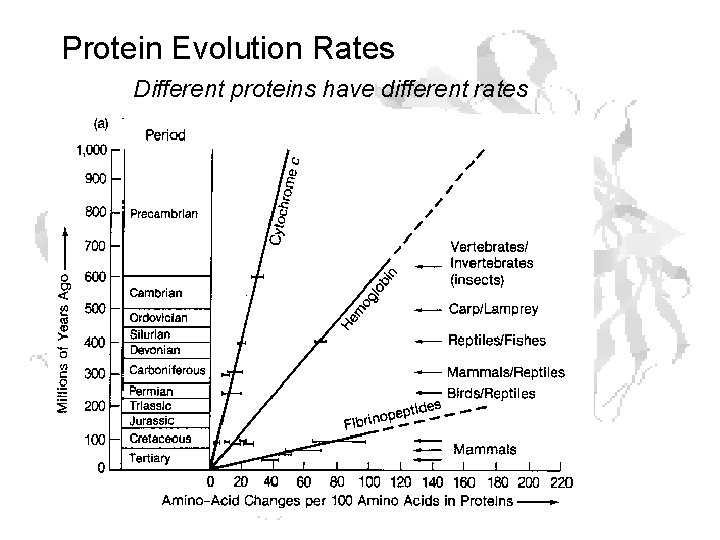
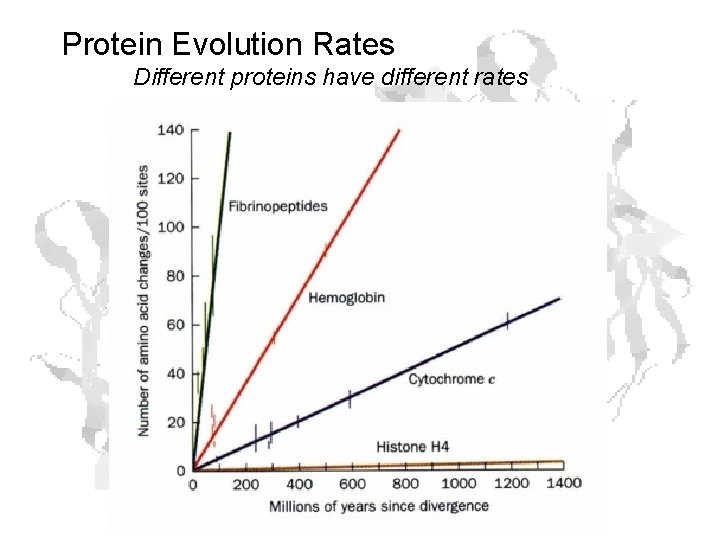
Protein Evolution Rates Different proteins have different rates

Protein Evolution Rates Different proteins have different rates

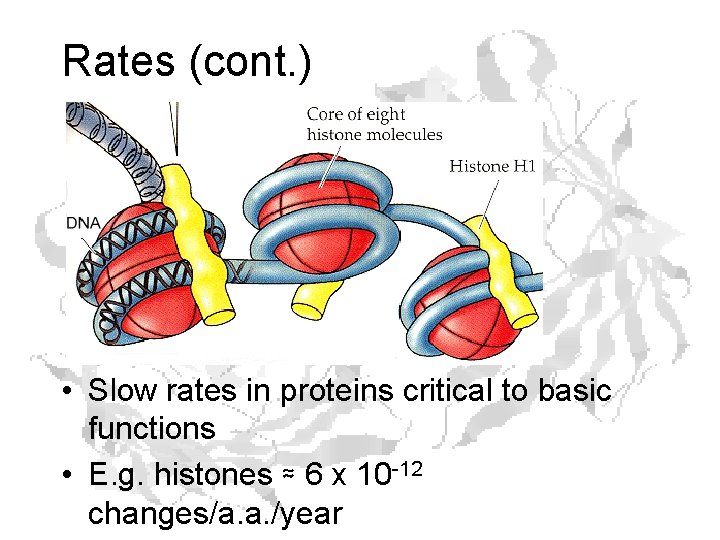
Rates (cont. ) • Slow rates in proteins critical to basic functions • E. g. histones ≈ 6 x 10 -12 changes/a. a. /year

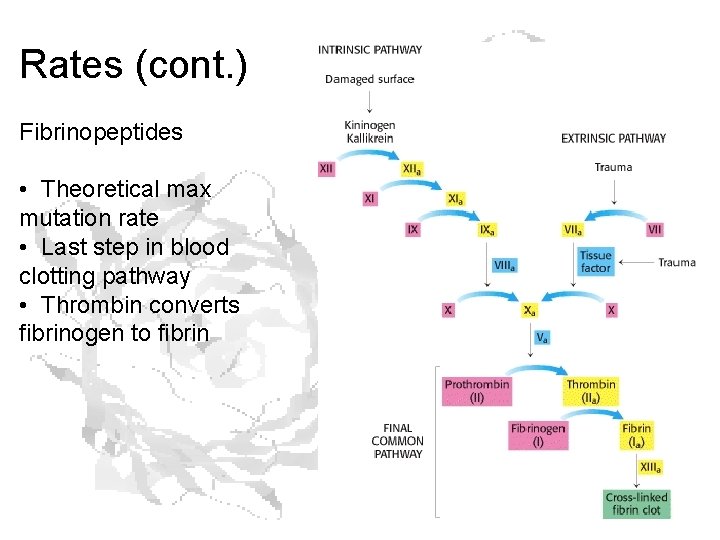
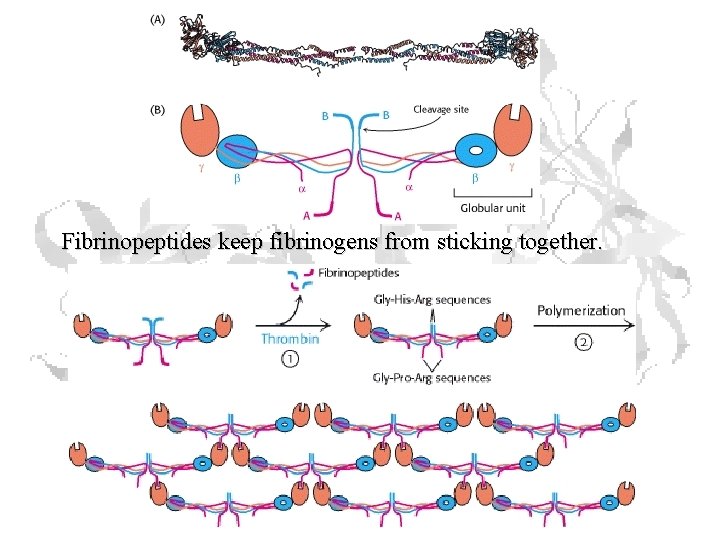
Rates (cont. ) Fibrinopeptides • Theoretical max mutation rate • Last step in blood clotting pathway • Thrombin converts fibrinogen to fibrin

Fibrinopeptides keep fibrinogens from sticking together.

Rates (cont. ) • Only constraint on sequence is that it has to physically be there • Fibrinopeptide limit ≈ 9 x 10 -9 changes/a. a. /year

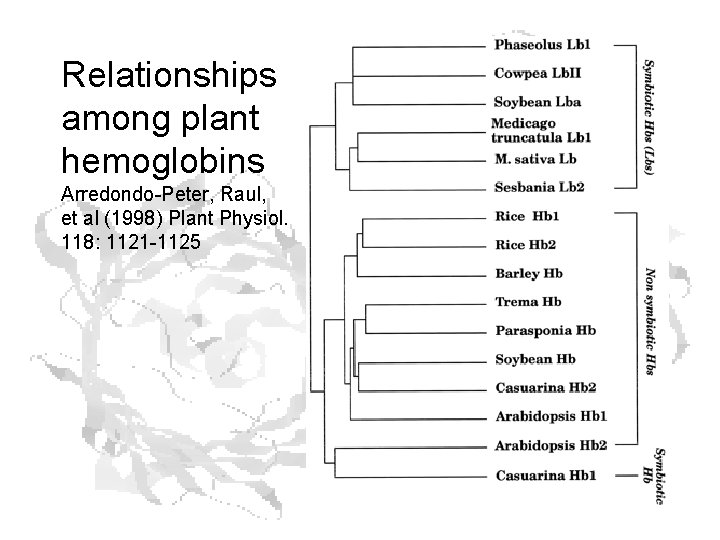
Relationships among plant hemoglobins Arredondo-Peter, Raul, et al (1998) Plant Physiol. 118: 1121 -1125

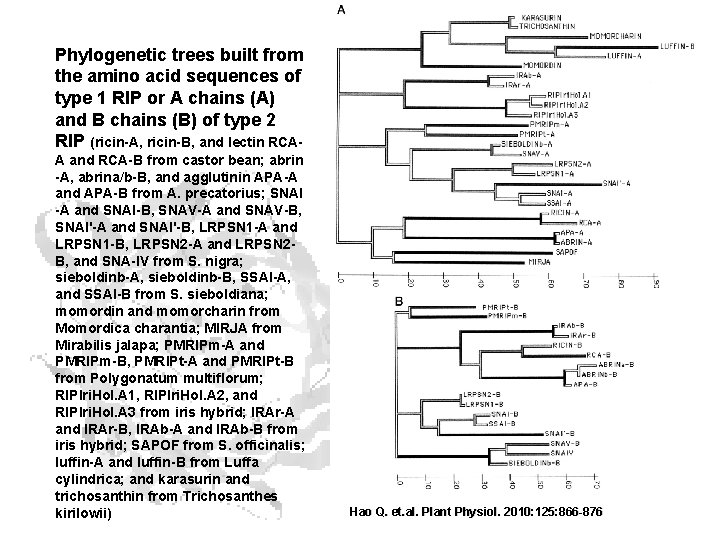
Amino acid sequences of several ribosome-inhibiting proteins

Phylogenetic trees built from the amino acid sequences of type 1 RIP or A chains (A) and B chains (B) of type 2 RIP (ricin-A, ricin-B, and lectin RCAA and RCA-B from castor bean; abrin -A, abrina/b-B, and agglutinin APA-A and APA-B from A. precatorius; SNAI -A and SNAI-B, SNAV-A and SNAV-B, SNAI'-A and SNAI'-B, LRPSN 1 -A and LRPSN 1 -B, LRPSN 2 -A and LRPSN 2 B, and SNA-IV from S. nigra; sieboldinb-A, sieboldinb-B, SSAI-A, and SSAI-B from S. sieboldiana; momordin and momorcharin from Momordica charantia; MIRJA from Mirabilis jalapa; PMRIPm-A and PMRIPm-B, PMRIPt-A and PMRIPt-B from Polygonatum multiflorum; RIPIri. Hol. A 1, RIPIri. Hol. A 2, and RIPIri. Hol. A 3 from iris hybrid; IRAr-A and IRAr-B, IRAb-A and IRAb-B from iris hybrid; SAPOF from S. officinalis; luffin-A and luffin-B from Luffa cylindrica; and karasurin and trichosanthin from Trichosanthes kirilowii) Hao Q. et. al. Plant Physiol. 2010: 125: 866 -876

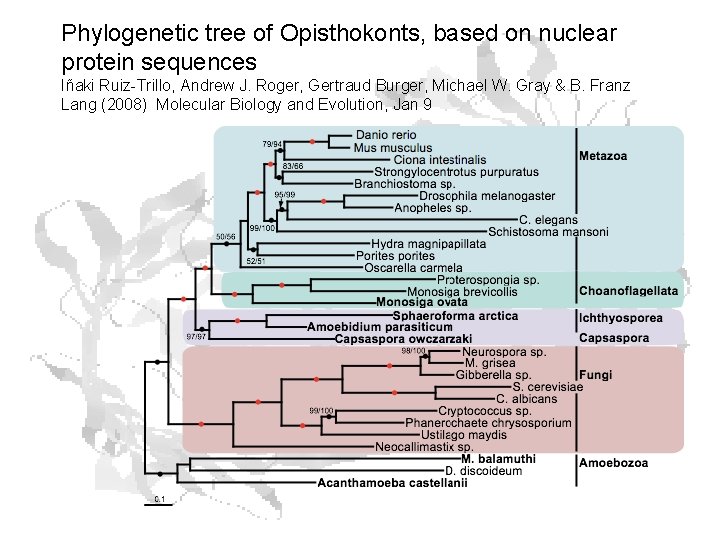
Phylogenetic tree of Opisthokonts, based on nuclear protein sequences Iñaki Ruiz-Trillo, Andrew J. Roger, Gertraud Burger, Michael W. Gray & B. Franz Lang (2008) Molecular Biology and Evolution, Jan 9