CFX 96 RealTime PCR Detection System Fast Friendly



























































- Slides: 62

CFX 96 Real-Time PCR Detection System Fast, Friendly, Flexible Designed for the Way You Work Rethink PCR Version 1. 0

Discussion for today AMPLIFICATION • Real time PCR technology • CFX 96 system features – Methods for optimization – Data Analysis • CFX 96 software Version 1. 0 www. bio-rad. com/pcr

What is Real-Time q. PCR? AMPLIFICATION • Fluorescence-based detection of amplification products through the use of a DNA-binding dye or hybridization probe. • Real-time q. PCR is used to quantify input nucleic acid by measuring the number of cycles required to reach a set level of product. • In contrast, traditional PCR is used to amplify DNA with end point analysis to distinguish products. Version 1. 0 www. bio-rad. com/pcr

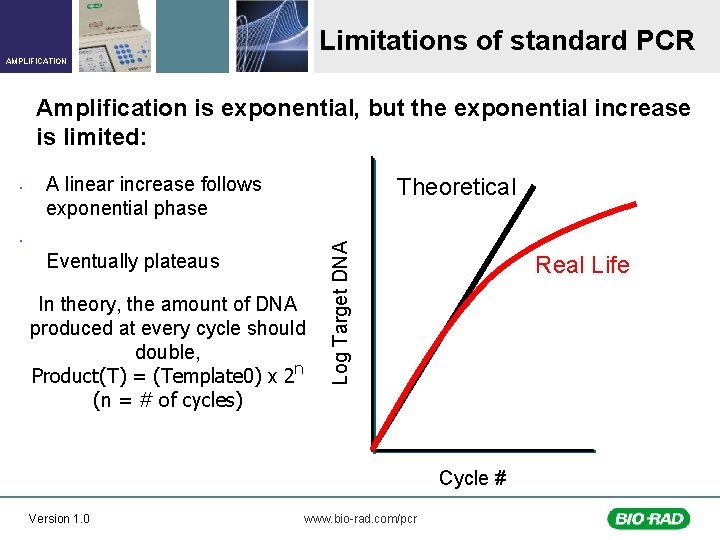
Limitations of standard PCR AMPLIFICATION Amplification is exponential, but the exponential increase is limited: A linear increase follows exponential phase Theoretical • Eventually plateaus In theory, the amount of DNA produced at every cycle should double, Product(T) = (Template 0) x 2 n (n = # of cycles) Log Target DNA • Real Life Cycle # Version 1. 0 www. bio-rad. com/pcr

AMPLIFICATION Standard PCR is as endpoint 96 identical reactions will have very different final amounts of fluorescence at endpoint Version 1. 0 www. bio-rad. com/pcr

AMPLIFICATION Real-Time PCR Through the use of fluorescent molecules, real-time PCR has the ability to directly measure the reaction while amplification is taking place. Version 1. 0 www. bio-rad. com/pcr

AMPLIFICATION How is quantitative data collected? Log Target DNA Theoretical Detector Cycle # Version 1. 0 Real Life www. bio-rad. com/pcr

AMPLIFICATION Threshold Cycle, CT 96 identical reactions will have almost identical CT values Version 1. 0 www. bio-rad. com/pcr

AMPLIFICATION Threshold Cycle, CT The point at which the fluorescence rises appreciably above background Threshold can be placed anywhere in the exponential (log-linear) phase Version 1. 0 www. bio-rad. com/pcr

Threshold Setting AMPLIFICATION • After baseline subtraction, a threshold line is set empirically or by a statistical calculation at a fluorescence value above background. Threshold Log View Version 1. 0 www. bio-rad. com/pcr

Mathematical Implications AMPLIFICATION Ideal PCR Product. T=(Template 0)2 Where n=Number of Cycles • 1 CT Difference = 2 fold difference in starting template amount • 3. 3 CT Difference = 10 fold difference in starting template amount Version 1. 0 www. bio-rad. com/pcr n

AMPLIFICATION • Threshold Cycle, CT Correlates strongly with the starting copy number Version 1. 0 www. bio-rad. com/pcr

Threshold Cycle, CT AMPLIFICATION • Correlates strongly with the starting copy number 2 n = 10 fold n ln 2 = ln 10 n = ln 10 ln 2 n = 3. 32 Version 1. 0 www. bio-rad. com/pcr

Real-Time PCR: Applications AMPLIFICATION Real-Time reaction monitoring provides information for relative or absolute measurements of starting material. • Gene Expression Studies • Chromatin Immunoprecipitation (Ch. IP) • Methylation Specific PCR (HRM) • Microarray Validation • Transgenic Analysis • GMO Testing • Viral/Bacterial Load Studies • Allelic Discrimination/SNP (HRM) Version 1. 0 www. bio-rad. com/pcr

AMPLIFICATION Version 1. 0 From CT values, we can determine the initial copy number www. bio-rad. com/pcr

AMPLIFICATION Chemistries used in real time PCR • Intercalation Dyes • Hybridization Probes Version 1. 0 www. bio-rad. com/pcr

AMPLIFICATION Intercalation (DNA binding) dyes n n n DNA binding dyes are inexpensive compared to hybridization probes. Et. Br is 25 times more fluorescent when bound to ds. DNA SYBR Green I is 125 times more fluorescent brightly bound to ds. DNA Version 1. 0 www. bio-rad. com/pcr

Intercalation Dyes: AMPLIFICATION SYBR Green I l l 3’ 5’ l ID ID ID l l Version 1. 0 5’ 5’ ID Taq ID www. bio-rad. com/pcr 3’

SYBR Green I AMPLIFICATION • Advantages – Experiment only requires primers • Disadvantages – Potential contribution to fluorescence from nonspecific products (primer-dimers) – No multiplexing Version 1. 0 www. bio-rad. com/pcr

Hybridization Probes AMPLIFICATION Currently, hybridization probe strategies fall into three main categories: • Cleavage-based assay • Taq. Manä Assays • Locked nucleic acids (LNA) • Displaceable probe assays • molecular beacons • Dual-oligo FRET probes • Probes incorporated directly into the primers • Amplifluor & Scorpions Version 1. 0 www. bio-rad. com/pcr

Cleavage-based assay: Taq. Man. TM AMPLIFICATION 5’ 3’ 3’ 3’ Add i. Q Supermix, Hybridization Probe and sample 5’ 5’ 3’ Primers 5’ 5’ Thermal Stable DNA Polymerase 3’ 3’ 5’ d. NTPs 3’ R 5’ 3’ 5’ 5’ Probe Q 3’ Denaturation 3’ 5’ Taq 3’ 3’ 3’ 5’ 5’ 3’ 5’ l 5’ Annealing Version 1. 0 www. bio-rad. com/pcr R 5’ Q 3’

Cleavage-based assay: Taq. Man. TM AMPLIFICATION Q R 5’ 3’ 3’ 5’ 5’ 3’ R Extension Step Q Taq 3’ 5’ 5’ 3’ R Q 3’ Taq 5’ 5’ 3’ R Taq Q 5’ 3’ l R Taq Q 3’ 5’ Version 1. 0 5’ 3’ www. bio-rad. com/pcr

Taq. Man AMPLIFICATION • Advantages – Target specific fluorescence – Multiplexing • Disadvantages – High initial cost – Assay design not trivial Version 1. 0 www. bio-rad. com/pcr

Real Time PCR Technology: AMPLIFICATION Real-Time PCR: -Enables detection and quantification of sample -Extremely sensitive -Can be used in various applications (gene expression, allelic discrimination, pathogen detection) Questions? Version 1. 0 www. bio-rad. com/pcr

AMPLIFICATION CFX 96 Real-Time PCR Detection System • Modular thermal cycler platform, includes C 1000 thermal cycler chassis, CFX 96 optical reaction module, CFX Manager software Version 1. 0 www. bio-rad. com/pcr

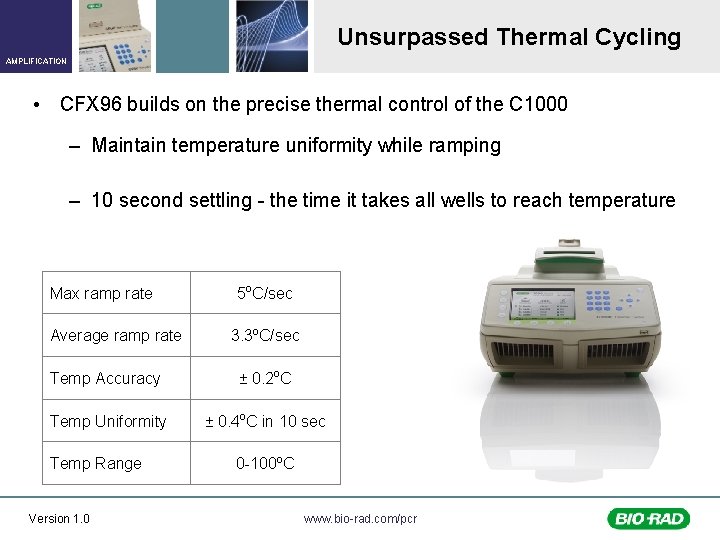
Unsurpassed Thermal Cycling AMPLIFICATION • CFX 96 builds on the precise thermal control of the C 1000 – Maintain temperature uniformity while ramping – 10 second settling - the time it takes all wells to reach temperature Max ramp rate Average ramp rate 5 o. C/sec 3. 3 o. C/sec Temp Accuracy ± 0. 2 o. C Temp Uniformity ± 0. 4 o. C in 10 sec Temp Range Version 1. 0 0 -100 o. C www. bio-rad. com/pcr

Patented Block Design AMPLIFICATION Fast block architecture Mass-reduced sample block* * Patented by Bio-Rad Version 1. 0 www. bio-rad. com/pcr

Time to Temperature AMPLIFICATION 1000 -Series Thermal Cycler Time to Temperature Probe Location Uniform ramping + shorter settling times = Faster PCR Version 1. 0 www. bio-rad. com/pcr

CFX 96 Optical Technology AMPLIFICATION • Scanning optics shuttle • 6 filtered LEDs for excitation • 6 filtered photodiodes for detection • Multiplex up to 5 targets • Independently illuminate and detect fluorescence in each channel during scan Version 1. 0 www. bio-rad. com/pcr

Next Generation Optical Technology: AMPLIFICATION • CFX 96 uses a scanning shuttle – 6 filtered LEDs for excitation – 6 filtered photodiodes for detection – LEDs fire sequentially • Multiplex up to 5 targets • All dyes excited near their maxima • Fixed optical path for all wells • No cross talk • Data is collected for all wells in all channels Version 1. 0 www. bio-rad. com/pcr

AMPLIFICATION Optical Technology provides hassle free maintenance • LEDs are long lasting • Factory calibrated. Does not require recalibration • No need for Passive Reference (Rox) • Data is always acquired from all wells in all channels • >100/well/scan • Laser Homing of shuttle at every scan Version 1. 0 www. bio-rad. com/pcr

Multiple Fast Scan Modes AMPLIFICATION Mode Channel(s) Scan Time (sec) All Channels 1 -5 12 SYBR/FAM Only 1 3 FRET 6 3 Version 1. 0 www. bio-rad. com/pcr

Excellent Uniformity at 10 l AMPLIFICATION Fast Scan All Channels Ave Ct = 19. 29 ± 0. 12 Ave Ct = 19. 81 ± 0. 12 Version 1. 0 www. bio-rad. com/pcr

Flexibility to use 6 Filter Sets AMPLIFICATION Channel Excitation (nm) Detection (nm) Calibrated Fluorophores 1 450 -490 515 -530 FAM™, SYBR Green I™ 2 515 -535 560 -580 VIC®, HEX™, TET™, Cal Gold 540™ 3 560 -590 610 -650 ROX™, TEXAS RED®, Cal Red 610™ 4 620 -650 675 -690 CY 5, Quasar 670™ 5 672 -684 705 -730 Quasar 705™ 6 450 -490 560 -580 Accommodates FRET Chemistry No need to recalibrate, ever. Reliable. Stable. Long life. Hassle free. Version 1. 0 www. bio-rad. com/pcr

Unsurpassed Dye Separation AMPLIFICATION Achieve sensitive multiplexing by maximal excitation and detection of dyes 60000 50000 40000 Signal 30000 20000 Q 705 10000 Cy 5 Tx. Red 0 1 Hex 2 Fam 3 4 Channel Version 1. 0 5 www. bio-rad. com/pcr Fluorophore

Excellent Uniformity at 10 l AMPLIFICATION Version 1. 0 Hex Texas Red Ave Ct = 19. 67 ± 0. 11 Max-Min =0. 52 Ave Ct = 19. 21 ± 0. 11 Max-Min =0. 61 www. bio-rad. com/pcr

Excellent Uniformity at 10 l in all channels AMPLIFICATION Version 1. 0 Cy 5 � Quasar 705 Ave Ct = 19. 96 ± 0. 12 Max-Min =0. 62 Ave Ct = 19. 27 ± 0. 07 Max-Min =0. 37 www. bio-rad. com/pcr

CFX 96 features for Reaction Optimization AMPLIFICATION • Melt Curve –MIQE Guidelines • Thermal Gradient • Fast RT-PCR • Data Analysis Version 1. 0 www. bio-rad. com/pcr

Melt Curve Analysis AMPLIFICATION • Principle: – After PCR amplification, the temperature is increased, causing the ds. DNA to melt and release SGI, resulting in a decrease in fluorescence • Analogous to agarose gel analysis except Tm is used to distinguish products • Melting temperature (Tm) of ds. DNA – Temperature at which half the DNA is double stranded and half is single stranded – Depends on nucleotide content and length Version 1. 0 www. bio-rad. com/pcr

Melt Curve Analysis AMPLIFICATION • After real-time PCR amplification, a melt curve is performed in presence of a DNA binding “saturation dye” • Melting temperature (Tm) – DNA is half double and half single-stranded – Depends on nucleotide content and length Double Stranded DNA Single Stranded Tm Version 1. 0 www. bio-rad. com/pcr

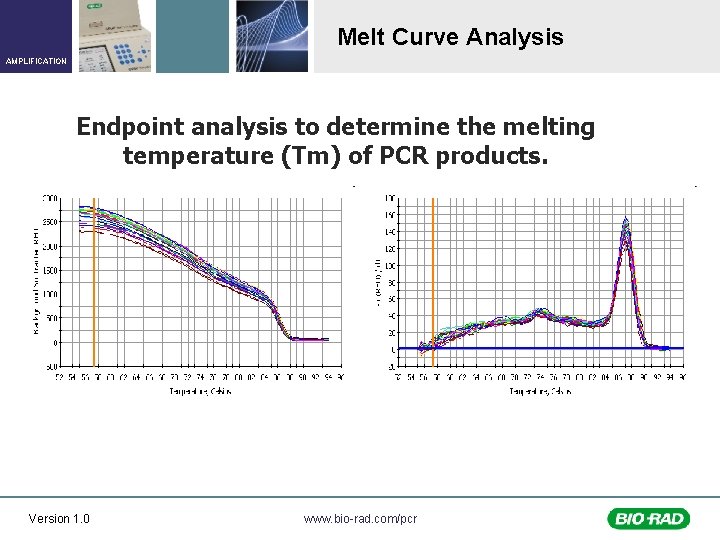
Melt Curve Analysis AMPLIFICATION Endpoint analysis to determine the melting temperature (Tm) of PCR products. Version 1. 0 www. bio-rad. com/pcr

AMPLIFICATION Version 1. 0 Melt Curve Analysis: Primer Dimer www. bio-rad. com/pcr

Thermal Gradient AMPLIFICATION • • • Version 1. 0 Used for one-step reactiontemperature optimization for PCR reaction specificity and efficiency. Up to 25 o. C gradient range programmable across block. “Dynamic Ramping” - cycler maintains the same hold time for each temperature. www. bio-rad. com/pcr

Thermal Gradient AMPLIFICATION 1) annealing temperature • Use temperature gradient feature 2) primer concentration • Look for lowest Ct value Dilution series of primer [ ] Temperature gradient Version 1. 0 SYBR Green I chemistry www. bio-rad. com/pcr

Optimization of Annealing Temperature for Best Results AMPLIFICATION • Annealing temperature is critical for Specificity Reproducibility PCR Reaction Efficiency Sensitivity Reliable data 67 o. C Efficiency = 68% • Serial dilutions 8 temps from 55 o. C to 68 o. C 62 o. C Efficiency = 99% 56 o. C Efficiency = 98% • 62 o. C is optimal -low Cts and highest reaction efficiency Version 1. 0 www. bio-rad. com/pcr

Fast PCR: 3 -step PCR vs 2 -step PCR AMPLIFICATION 95ºC 72ºC 58ºC Denaturation Version 1. 0 Annealing www. bio-rad. com/pcr Extension

AMPLIFICATION Version 1. 0 Quality Assays – Sso. Fast Eva Green Supermix www. bio-rad. com/pcr

Sso. Fast Eva Green Supermix: AMPLIFICATION Sso 7 d-fusion Protein Technology Sso 7 d from Sulfolobus solfataricus – – 7 k. D, 63 aa. Thermostable (Tm >90°C) No sequence preference Binds to ds. DNA (3 -6 bp/protein molecule) – Monomeric Version 1. 0 www. bio-rad. com/pcr

Sso. Fast Eva Green Supermix: Eva. Green Dye AMPLIFICATION • Eva. Green dye is similar to SYBR® Green I • Very low PCR inhibition • Increased sensitivity • Fast q. PCR Version 1. 0 www. bio-rad. com/pcr

Data Analysis: AMPLIFICATION • Basic delta Ct • Delta-delta Ct • Pfaffl delta-delta Ct Version 1. 0 www. bio-rad. com/pcr

Calculating for relative quantitation AMPLIFICATION Basic delta Ct method: (no normalization to reference gene) Primer set #2 22 24 Tissue #1: Tissue #2: Delta Ct: Fold induction = Version 1. 0 24 -22 = 2 22 = 4 www. bio-rad. com/pcr

Calculating for relative quantitation AMPLIFICATION Delta-delta Ct method: (assumes same efficiencies for each primer set) Reference Primer set Tissue #1: 21 Tissue #2: 1 st Delta 2 nd Delta 20 24 Delta Ct: 22 -21 = 1 Delta Ct: 24 -20 = 4 Delta Ct: 4 -1 = 3 Fold induction = 23 = 8 Version 1. 0 GOI Primer set 22 www. bio-rad. com/pcr

Calculating for relative quantitation AMPLIFICATION Problems of delta-delta Ct method: Ct 24 22 90% SQ Version 1. 0 www. bio-rad. com/pcr

Calculating for relative quantitation AMPLIFICATION Problems of delta-delta Ct method: Ct 24 22 90% 100% SQ Version 1. 0 www. bio-rad. com/pcr

Calculating for relative quantitation AMPLIFICATION Problem with the CT Slopes are not parallel Ct 24 22 90% 100% Starting quantity Version 1. 0 www. bio-rad. com/pcr

Calculating for relative quantitation AMPLIFICATION Pfaffl method: (Pfaffl, 2001; Nucleic Acid Research) Efficiencytarget delta. Ct target (control-sample) Fold induction = Efficiencyreference delta. Ct reference (control-sample) Efficiency = 10 -1/slope Version 1. 0 www. bio-rad. com/pcr

Calculating for relative quantitation AMPLIFICATION Pfaffl method: (efficiencies are normalized) Primer set #1 Reference (From Standard curve) Tissue #1: 21 22 Tissue #2: 20 24 Efficiency: 90% = 1. 9 Delta Ct: 20 -21 = -1 Fold induction = 2 target delta. Ct target (24 -22 1. 9 reference Version 1. 0 Primer set #2 GOI 24 -22 = 2) delta. Ct reference (20 -21 www. bio-rad. com/pcr 100% = 2 = -1) = 4 0. 53 = 7. 5

AMPLIFICATION Comparison of methods for relative quantitation calculations § Basic delta Ct method: (no reference gene) § Fold induction : 4 § Delta-delta Ct method: (reference gene) § Fold induction : 8 § Ideal for primer pairs with an E ≥ 90% AND large fold changes in expression (10 fold or more) § Pfaffl method: (reference gene and efficiency) § Fold induction : 7. 5 Version 1. 0 www. bio-rad. com/pcr

Relative Gene Expression Analysis AMPLIFICATION What to Use as Standards • Plasmid DNA • PCR Product • Spiked sample (with plasmid or PCR product) • Positive c. DNA control but unknown concentration (dilution) Version 1. 0 www. bio-rad. com/pcr

Vandesompele Method AMPLIFICATION • There are no true “House keeping” genes • Uses more than 1 reference gene (3 is recommended) and takes the geometric mean to normalize fold expression • Using a single reference gene leads to erroneous normalization up to 3. 0 -fold and 6. 4 -fold in 25% and 10% of the cases, respectively, with sporadic values above 20 -fold • ge. Norm site: http: //medgen. ugen. be/~jvdesomp/genorm/ – ge. Norm is a popular algorithm to determine the most stable reference (housekeeping) genes from a set of tested candidate reference genes in a given sample panel Version 1. 0 www. bio-rad. com/pcr

Bio-Rad: Experts in Real-time PCR AMPLIFICATION • Bio-Rad’s Innovation in Real-time PCR continues with the CFX 96 • We can help you achieve success at every step of your research – In-house Scientists – Field Application Scientists – Field Service – Technical Support – Field Sales Representatives – www. bio-rad. com/genomics Version 1. 0 www. bio-rad. com/pcr Rethink PCR

CFX 96 Real-Time PCR System AMPLIFICATION Questions? Thank you for joining us! Version 1. 0 www. bio-rad. com/pcr