Central Forensic Laboratory of the Police Headquarters Warsaw

Central Forensic Laboratory of the Police Headquarters, Warsaw, Poland Evaluation of the methods for DNA extraction from archival blood samples Igor Olewiecki M. Sc. Joanna Karolina Purzycka M. Sc. Ireneusz Soltyszewski Ph. D.

DNA forensic analysis 1. 2. 3. 4. 5. 6. Collection and examination of biological samples DNA extraction Quantification of isolated DNA amplification Capillary electrophoresis Genotyping

DNA source Ø Saliva Ø Blood Ø Tissue (i. e. muscle, liver, brain) Ø Bone marrow Ø Hair Ø Sperm

DNA extraction Goals: 1. isolation of nucleic acid from the cell 2. purification of isolated DNA Yield/quality ratio should be as high as possible

DNA extraction methods: Ø Organic Ø Ion-exchange Ø Silica Resin Matrix Ø High Salt Ø Sodium Hydroxide Ø Magnetic Affinity Resin

Organic Ø Lysis of the cells: alkaline buffer or chaotropic agents, proteinase K, incubation in 56ºC Ø Purification: phenol: chloroform Ø Extraction: ethanol or isopropanol
Ion-exchange Resin Ø Lysis of the cells: sample is heated in the presence of resin to 100ºC Ø Purification: resin binds cellular components other than DNA Ø Extraction: centrifugation removes resin leaving DNA in the supernatant

Silica matrix Ø Lysis of the cells: chaotropic agents, proteinase K, incubation in 56ºC Ø Purification: DNA binds to the silica matrix contaminants are eliminated by washing steps Ø Extraction: DNA is rinsed with water from the matrix

Why to use a commercial kits? Ø Non-toxic methods Ø Short incubation and centrifugation times Ø Easy procedure steps Ø Able to get rid of inhibitors that are solvable in the aqueous phase Ø Ready for automatisation
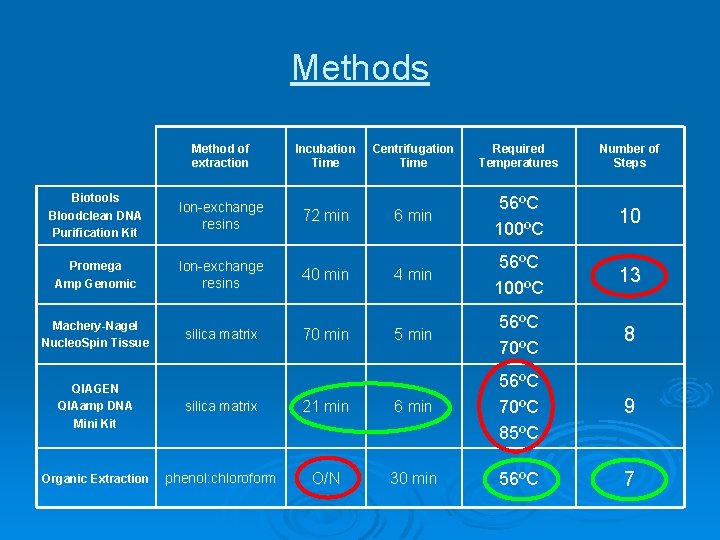
Methods Method of extraction Incubation Time Centrifugation Time Required Temperatures Number of Steps Biotools Bloodclean DNA Purification Kit Ion-exchange resins 72 min 6 min 56ºC 100ºC 10 Promega Amp Genomic Ion-exchange resins 40 min 4 min 56ºC 100ºC 13 Machery-Nagel Nucleo. Spin Tissue silica matrix 70 min 56ºC 70ºC 8 9 7 QIAGEN QIAamp DNA Mini Kit silica matrix 21 min 6 min 56ºC 70ºC 85ºC Organic Extraction phenol: chloroform O/N 30 min 56ºC
DNA measurement Ø DNA concentration was measured fluorometrically using Pico. Green ds. DNA Quantitation Reagent (Invitrogen Molecular Probes) and Fluoroscan Ascent FL (Labsystems) Ø Slot-blot technique specific to measure only the human DNA with Quantiblot kit (Applied Biosystems).
SGM plus analysis Ø 1, 25 µg of DNA in volume of 25 µl was taken for PCR reaction Ø PCR reaction was performed in a standard conditions Ø STR analysis was done with Gen. Scan 3. 7 and Genotyper 3. 7 software (default settings)

Material Ø Blood samples were collected in 1955 from 30 non-related individuals Ø Dried samples were packed in a paper envelopes, and stored at the room temperature and constant humidity Ø For DNA extraction 0, 02 g of dried blood was used from each sample
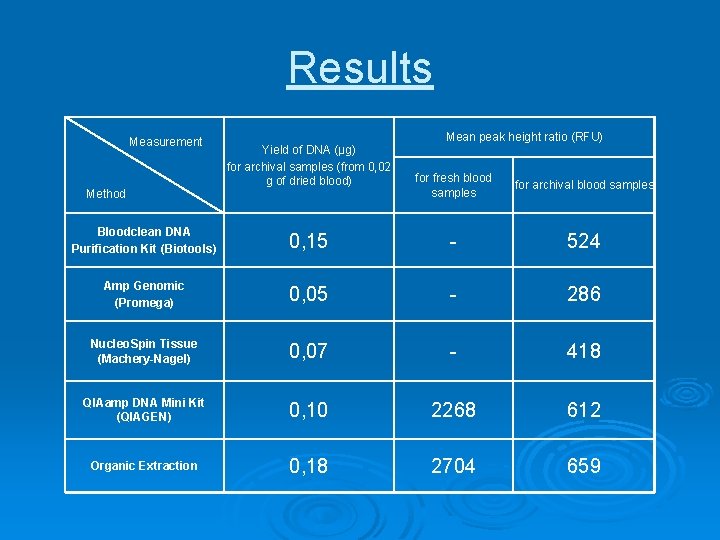
Results Measurement Method Yield of DNA (µg) for archival samples (from 0, 02 g of dried blood) Mean peak height ratio (RFU) for fresh blood samples for archival blood samples Bloodclean DNA Purification Kit (Biotools) 0, 15 - 524 Amp Genomic (Promega) 0, 05 - 286 Nucleo. Spin Tissue (Machery-Nagel) 0, 07 - 418 QIAamp DNA Mini Kit (QIAGEN) 0, 10 2268 612 Organic Extraction 0, 18 2704 659
Conclusion Ø It is possible to isolate DNA from archival blood samples that are almost 50 years old using different methods Ø It is possible to obtain a full genetic profile form such a DNA Ø QIAamp was the best commercial kit for DNA extraction from archival blood samples
- Slides: 15