Cellular clock mechanism First genetic results in mammals





- Slides: 11

Cellular clock mechanism

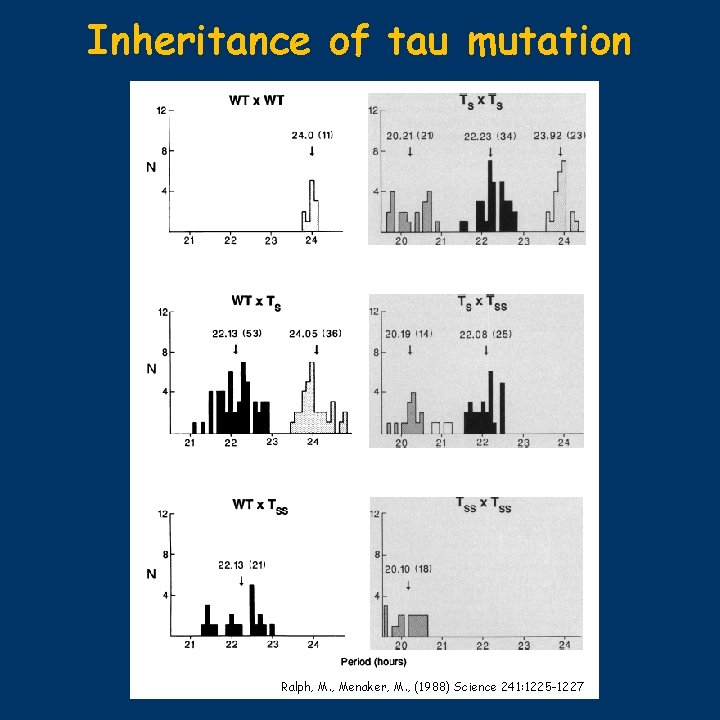
First genetic results in mammals • in 1985 Martin Ralph noticed a hamster that had an abnormally short period in DD • breeding the animal with normal females proved that the aberrant period length followed Mendelian intermediary inheritance: 24/22/20 for homo- and heterozygotes • the “founding father” was released by animal rights people from the lab… • the mutated gene was named tau, and it was the first example in vertebrates for a single gene influencing period length – in Drosophila and Neurospora such mutants were known • in SCN transplantation experiments, donor SCN was taken from homozygote tau-mutants – it proved that the recipient’s rhythms are controlled by the transplanted SCN 2/9

Further development 3/9 • forward genetic method means induction of random mutations by chemicals or irradiation hoping for a useful one • it was successfully applied in Drosophila (per), Neurospora (frq) and unicellular organisms, revealing genes related to circadian rhythms • Joseph Takahashi planned to use it in mammals despite many objections: – it was supposed that several genes participate in such a complex behavior and mutation in one of them will not have a strong enough impact – coexistence of several mutations will make analysis difficult or impossible • a young Ph. D student, Martha Vitaterna was assigned to the task, and she was lucky enough in 1994 to find a mutant in the first 42 mice • the mutation caused extremely long (28 h) period, and abolished persistence of rhythms

Finding the first clock gene 4/9 • the mutant gene was named Clock (circadian locomotor output cycles kaput) • genetics of mice was better known in 1994, thus the gene product was identified • it is a transcriptional factor carrying a basic loop -helix-loop (b. HLH) type DNA-binding domain, a PAS dimerization domain, and a glu-rich activation domain • PAS comes from Drosophyla PER (Period), human ARNT (Aryl hydrocarbon receptor translocator), Drosophyla SIM (Single minded) • in the mutant there is a point mutation (A T) causing the loss of a glu-rich exon important for the activation of transcription • presence of the PAS domain indicated that CLOCK formed a dimer with another PAS containing peptide to activate transcription

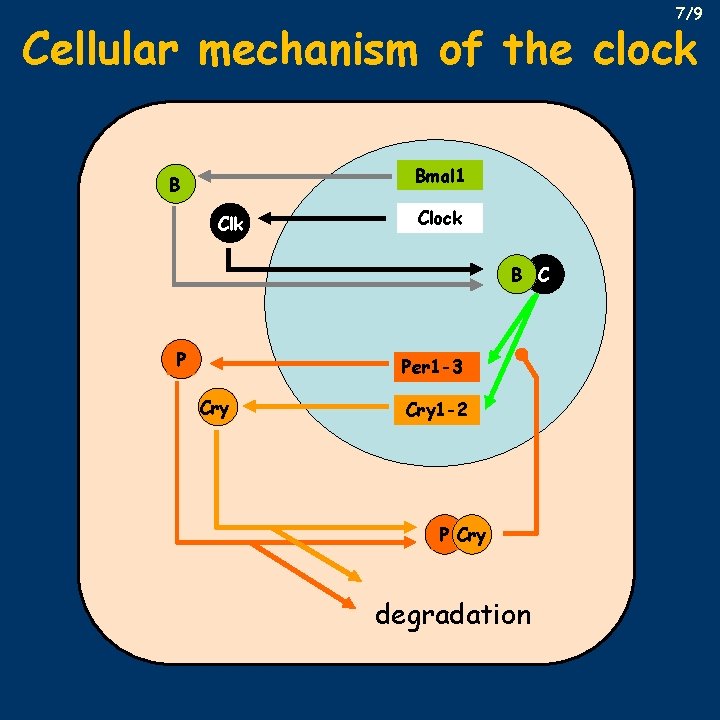
Clock genes 5/9 • by searching for PAS containing proteins that are able to dimerize with CLOCK, BMAL 1 (Brain and Muscle ARNT-Like 1) was found • CLOCK: BMAL 1 heterodimer was shown to regulate, through a so-called E-box (CACGTG), transcription of Per 1 -3 genes originally identified in Drosophila • this discovery led to the identification of Cry 12 (cryptochrome genes) that are also regulated through the E-box, and their products are able to form homodimers, or heterodimers with PER to inhibit CLOCK: BMAL 1 effect • of the Casein Kinase 1 (CK 1) family, ε and δ phosphorylates PER, promoting its degradation – tau mutant has a defective CK 1ε • several other genes have been identified that participate in the functioning of the mammalian clock

Conclusions about clock genes 6/9 • there are differences between the different species (Neurospora, Drosophila, mammals, etc. ), but the basics are the same • E-box can regulate many genes – clock regulated genes • clock genes are present in every cell, they can be transcribed and translated – peripheral clocks can be present in any cell • it explains the persistence of rhythms in isolated organs, dissociated cells, etc. , and also the desynchronization of physiological rhythms • individual cells are able to produce circadian signals, but how they are synchronized is still unknown • temperature independency of the clock can be explained by diffusion steps and by enzymatic reactions working against each other

7/9 Cellular mechanism of the clock Bmal 1 B Clk Clock B C P Per 1 -3 Cry 1 -2 P Cry degradation

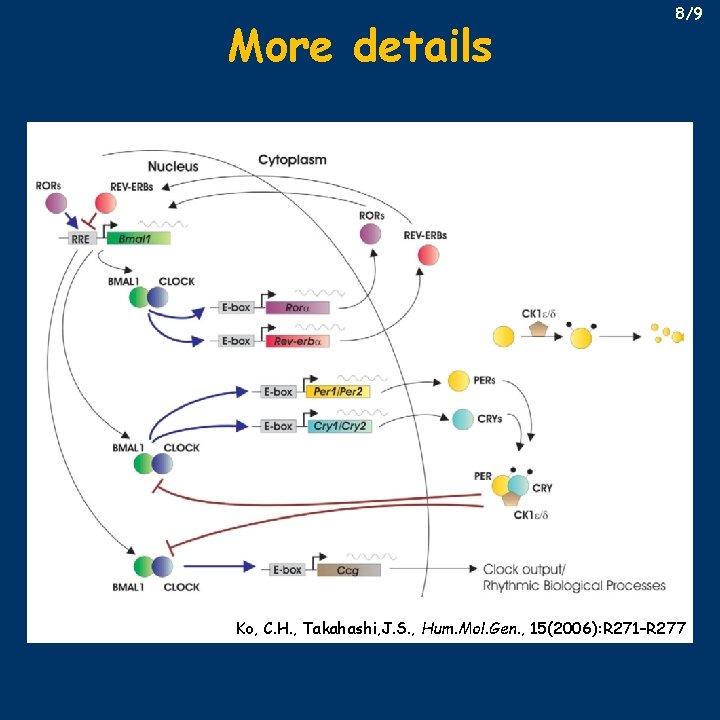
More details 8/9 Ko, C. H. , Takahashi, J. S. , Hum. Mol. Gen. , 15(2006): R 271–R 277

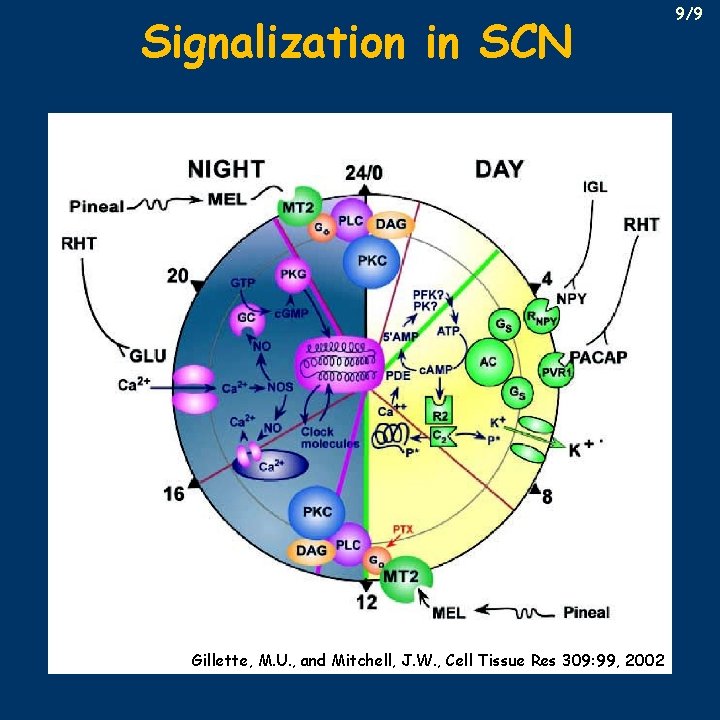
Signalization in SCN Gillette, M. U. , and Mitchell, J. W. , Cell Tissue Res 309: 99, 2002 9/9


Inheritance of tau mutation Ralph, M. , Menaker, M. , (1988) Science 241: 1225 -1227