Cellerator A System for Simulating Biochemical Reaction Networks
Cellerator: A System for Simulating Biochemical Reaction Networks Bruce E Shapiro bshapiro@jpl. nasa. gov Jet Propulsion Laboratory California Institute of Technology
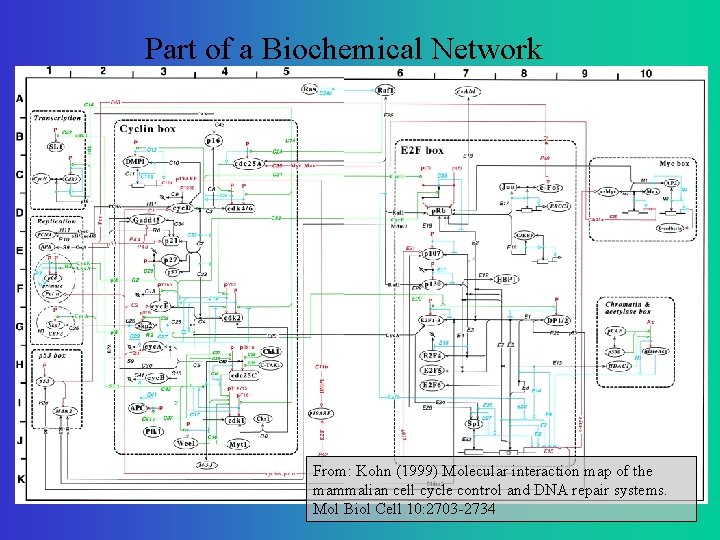
Part of a Biochemical Network From: Kohn (1999) Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Mol Biol Cell 10: 2703 -2734
Biochemical Networks Are. . . • Complex • Mutually interacting • Large – Number of reactions grows exponentially with number of states • Best understood pictorially • Best described quantitatively by a large system of differential equations (ODEs) Need to translate pictures to ODEs
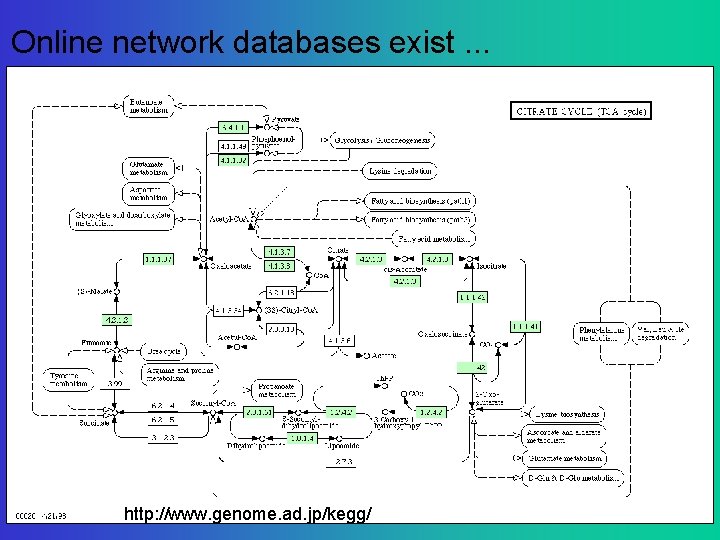
Online network databases exist. . . http: //www. genome. ad. jp/kegg/
. . . but mathematical simulations of these networks are hopelessly naive. . .
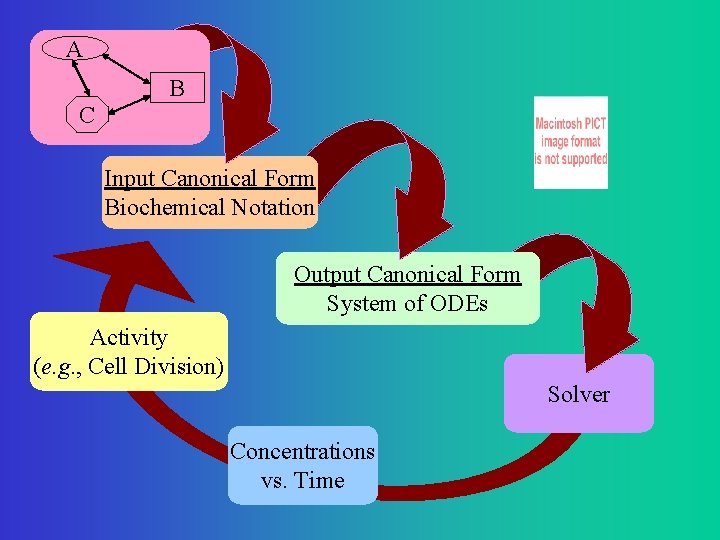
A B C Input Canonical Form Biochemical Notation Output Canonical Form System of ODEs Activity (e. g. , Cell Division) Solver Concentrations vs. Time
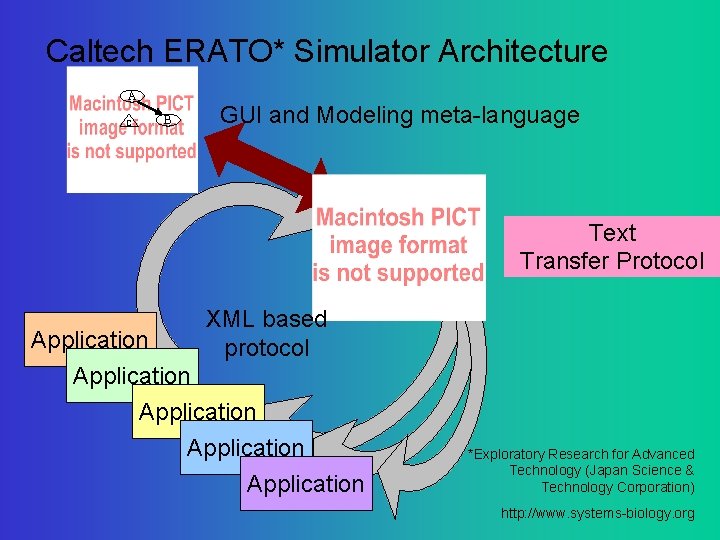
Caltech ERATO* Simulator Architecture A C B GUI and Modeling meta-language Text Transfer Protocol XML based protocol Application Application *Exploratory Research for Advanced Technology (Japan Science & Technology Corporation) http: //www. systems-biology. org
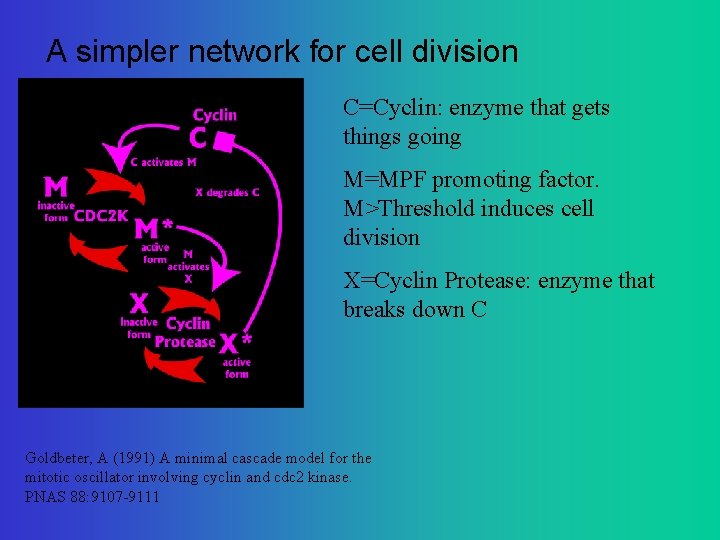
A simpler network for cell division C=Cyclin: enzyme that gets things going M=MPF promoting factor. M>Threshold induces cell division X=Cyclin Protease: enzyme that breaks down C Goldbeter, A (1991) A minimal cascade model for the mitotic oscillator involving cyclin and cdc 2 kinase. PNAS 88: 9107 -9111
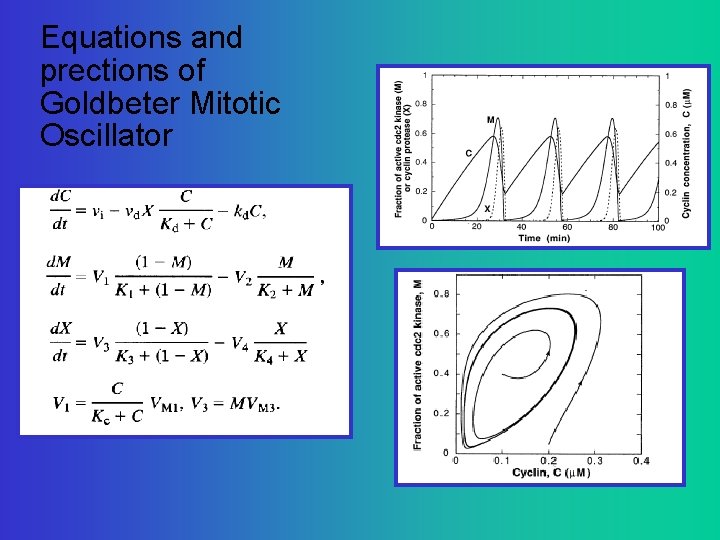
Equations and prections of Goldbeter Mitotic Oscillator
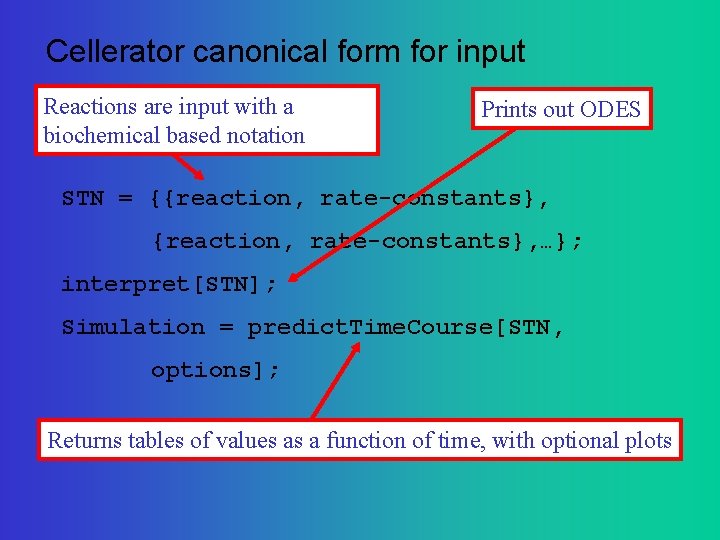
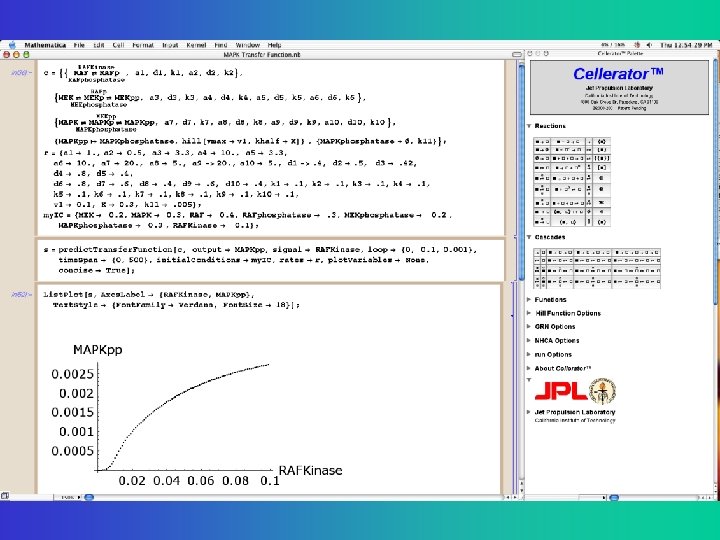
Cellerator canonical form for input Reactions are input with a biochemical based notation Prints out ODES STN = {{reaction, rate-constants}, …}; interpret[STN]; Simulation = predict. Time. Course[STN, options]; Returns tables of values as a function of time, with optional plots
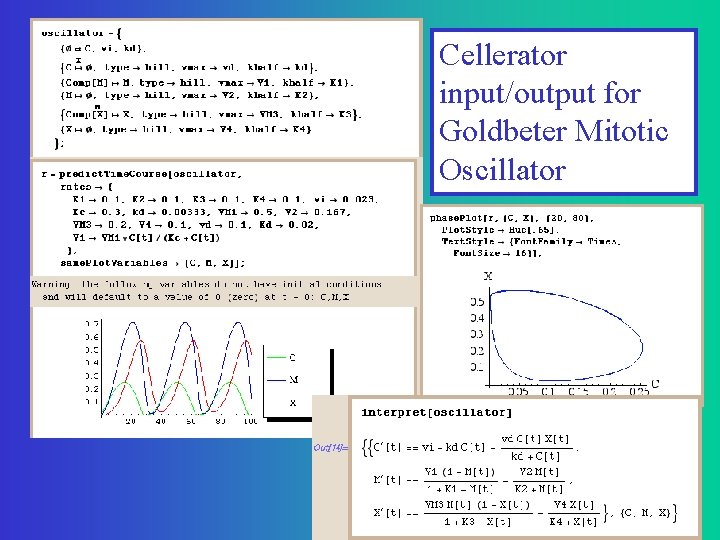
Cellerator input/output for Goldbeter Mitotic Oscillator
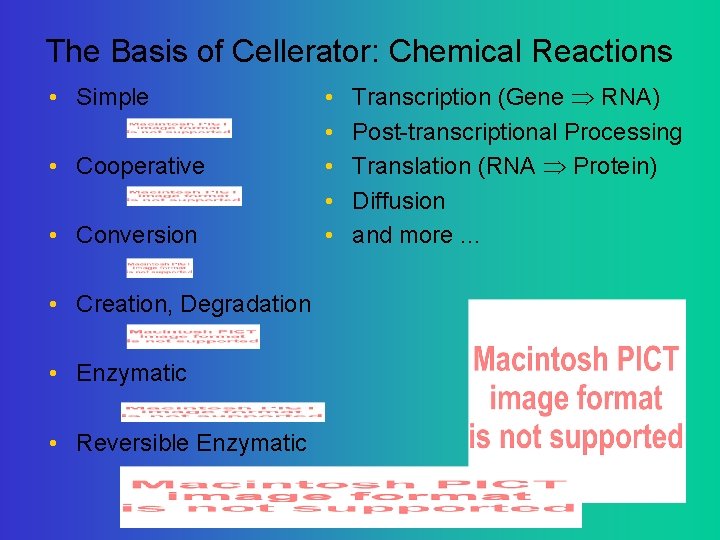
The Basis of Cellerator: Chemical Reactions • Simple • Cooperative • Conversion • Creation, Degradation • Enzymatic • Reversible Enzymatic • • • Transcription (Gene RNA) Post-transcriptional Processing Translation (RNA Protein) Diffusion and more. . .
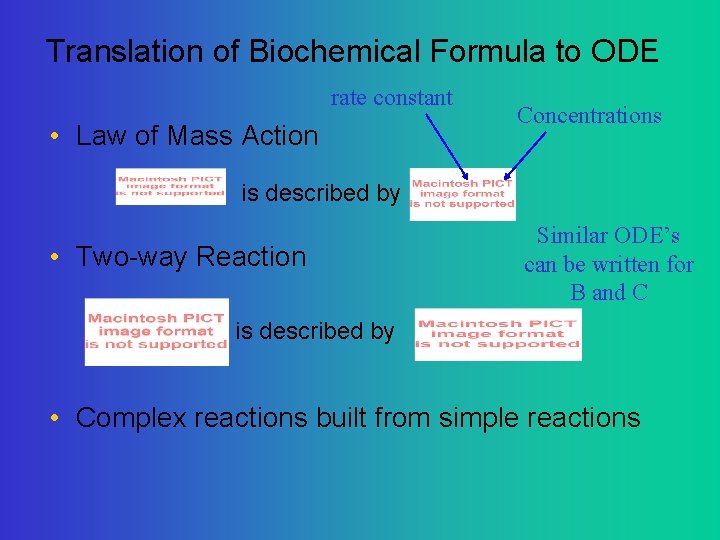
Translation of Biochemical Formula to ODE rate constant • Law of Mass Action Concentrations is described by • Two-way Reaction Similar ODE’s can be written for B and C is described by • Complex reactions built from simple reactions
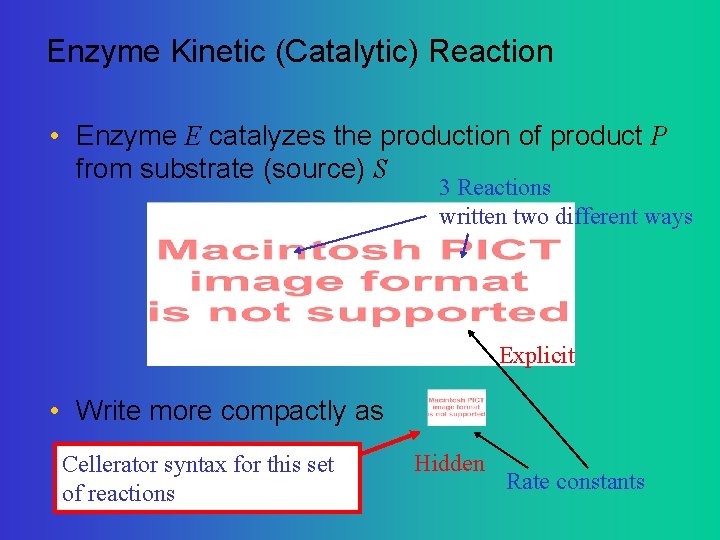
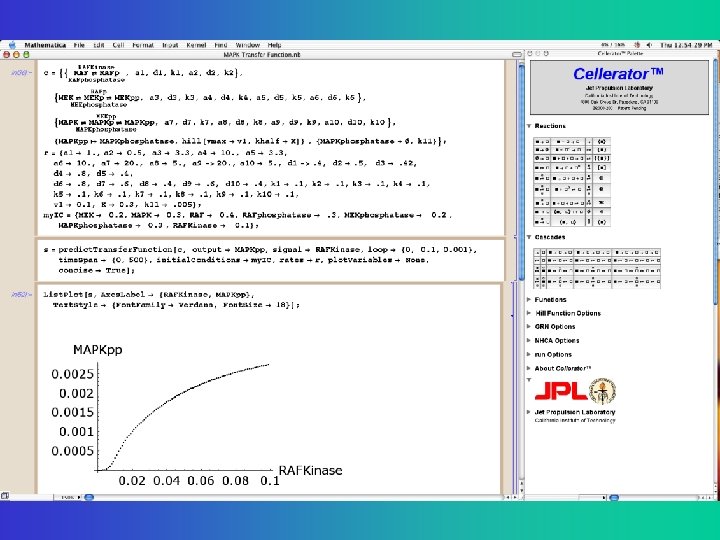
Enzyme Kinetic (Catalytic) Reaction • Enzyme E catalyzes the production of product P from substrate (source) S 3 Reactions written two different ways Explicit • Write more compactly as Cellerator syntax for this set of reactions Hidden Rate constants
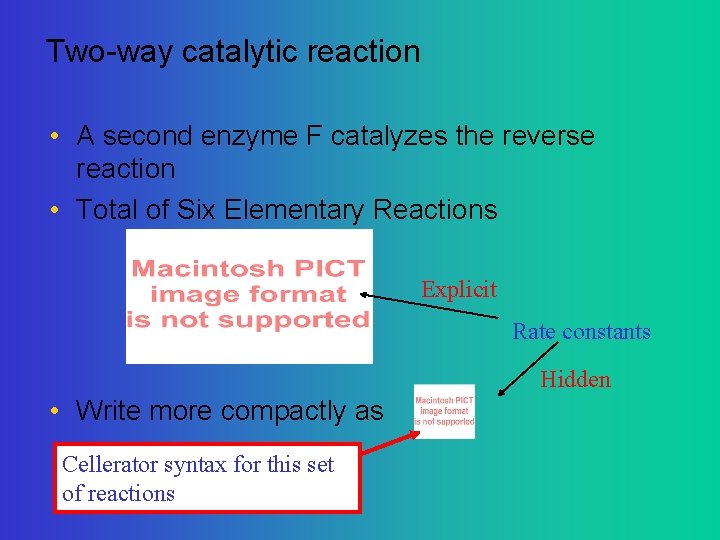
Two-way catalytic reaction • A second enzyme F catalyzes the reverse reaction • Total of Six Elementary Reactions Explicit Rate constants Hidden • Write more compactly as Cellerator syntax for this set of reactions

Canonical Forms for Translation: Chemical reactions • Input Canonical Form for Chemical Reaction • Output Canonical Form: Terms in an ODE

Cellerator Arrows: Law of Mass Action

Cellerator Arrows: Catalytic Reactions
Cellerator Arrows: Transcriptional Regulation
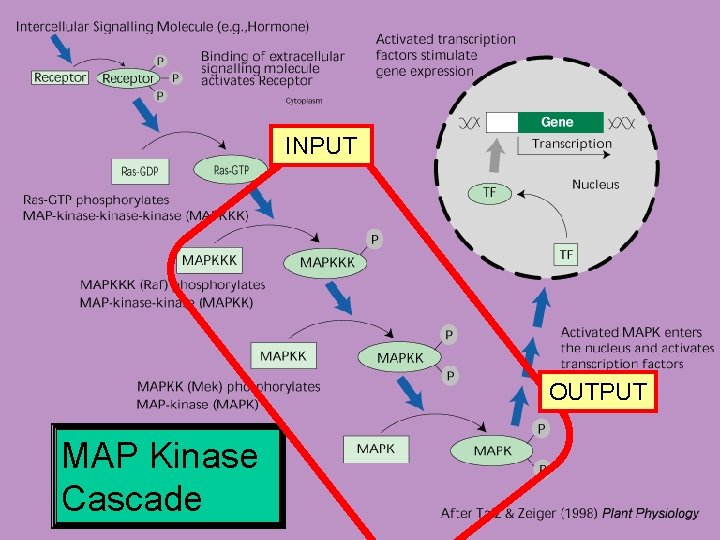
INPUT OUTPUT MAP Kinase Cascade

MAP Kinase in Scaffold
The combinatoric explosion
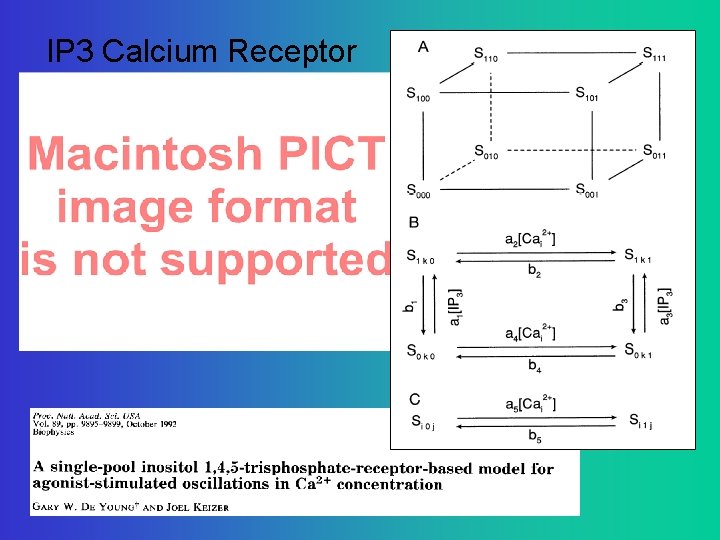
IP 3 Calcium Receptor
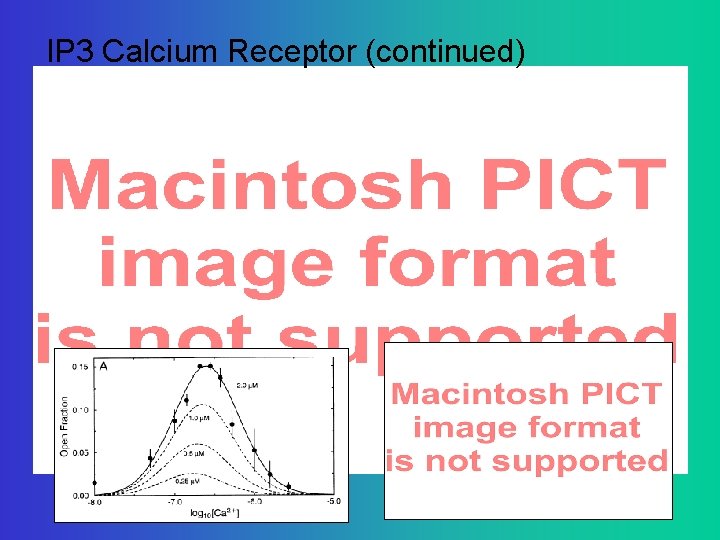
IP 3 Calcium Receptor (continued)
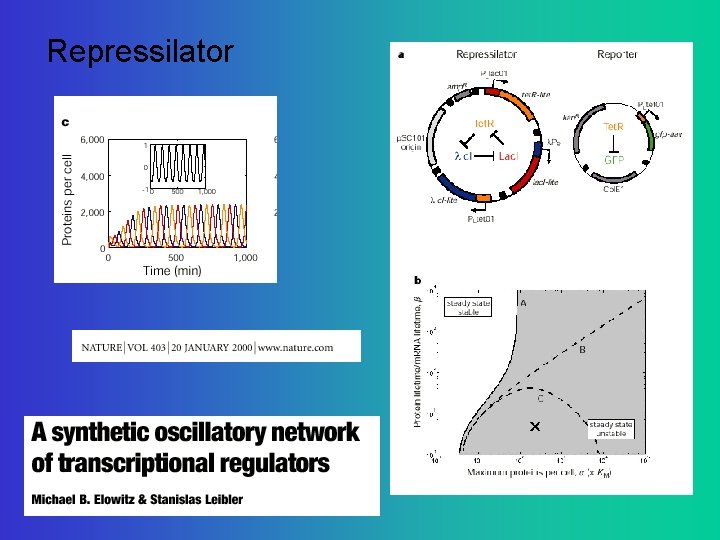
Repressilator

Repressilator
Object Oriented Implementation: “Domains” and “Fields” • • Domain: object Field: function that maps domains to R Field of Domains: maps domain elements to domains Example – graph. Domain: represents tissue – node Domains: cells – neighbors[g, n] returns a list of node. Domains that are neighbors of node n n in graph g

Multicellular Organisms
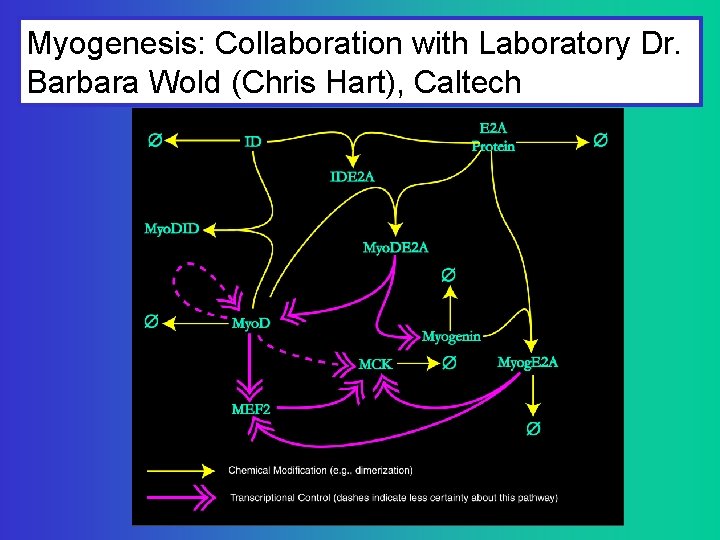
Myogenesis: Collaboration with Laboratory Dr. Barbara Wold (Chris Hart), Caltech
Plant Growth: Collaboration with Laboratory Dr. Elliot Meyerowitz, Caltech

Secondary Leukemia: Collaboration with City of Hope National Medical Center (NASA/BSRP) Focus: Pathogenesis of myelodysplasia & acute myeloid leukemia following high-dose chemo/radiotherapy and autologous peripheral blood stem cell transplantation for treatment of Hodgkin’s disease and non-Hodgkin’s lymphoma
JPL Collaborations using Cellerator • Effects of microgravity during space flight on bone and muscle development (Caltech, JSC, and UCI) • Development of childhood leukemias (Caltech, Children’s Hospital of LA, and UC, Irvine) • Description of “core” signal transduction units (Johns Hopikins) • Improving algorithms for micro-array data analysis (Caltech, Harvey Mudd) • Systems Biology Workbench (Caltech, JST/Erato)

Acknowledgements • • Eric Mjolsness* - UC, Irivine Andre Levchenko* - Johns Hopkins University Barbara Wold - Caltech Elliot Meyerowitz - Caltech * Original Developers
- Slides: 35