Cell structure Nucleoid Single strand of DNA usually


- Slides: 18

Cell structure

Nucleoid • Single strand of DNA, usually circular, usually looks like a big ball of messed up twine… • Size – smallest organism yet discovered (Nanoarchaeum equitans) 490, 889 base pairs; e. coli 4. 7 Mbp, most prokaryotes 1 -6 million base pairs (1 -6 MBp); Humans 3300 MBp • DNA is around 1000 mm long in bacteria, while the organism is on the order of 1 mm long – special enzymes called gyrases help coil it into a compact form

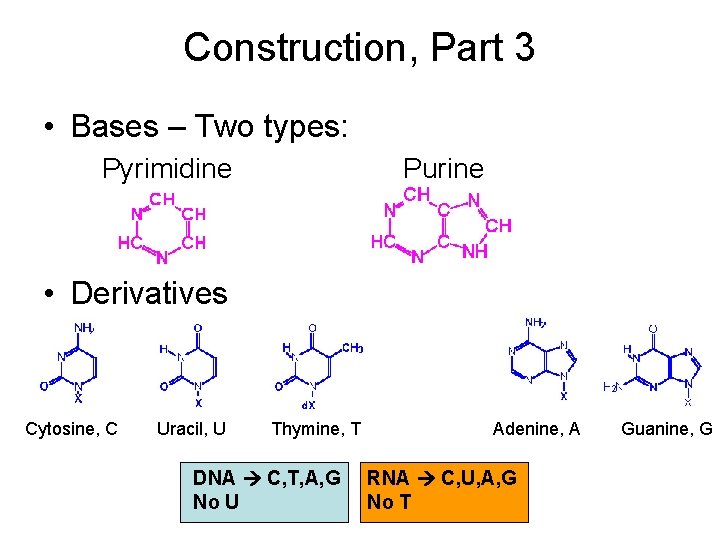
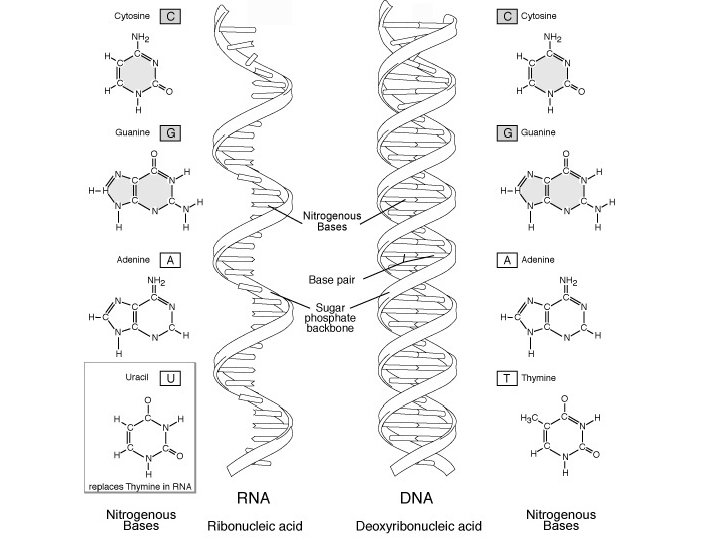
Construction, Part 3 • Bases – Two types: Pyrimidine Purine • Derivatives Cytosine, C Uracil, U Thymine, T DNA C, T, A, G No U Adenine, A RNA C, U, A, G No T Guanine, G

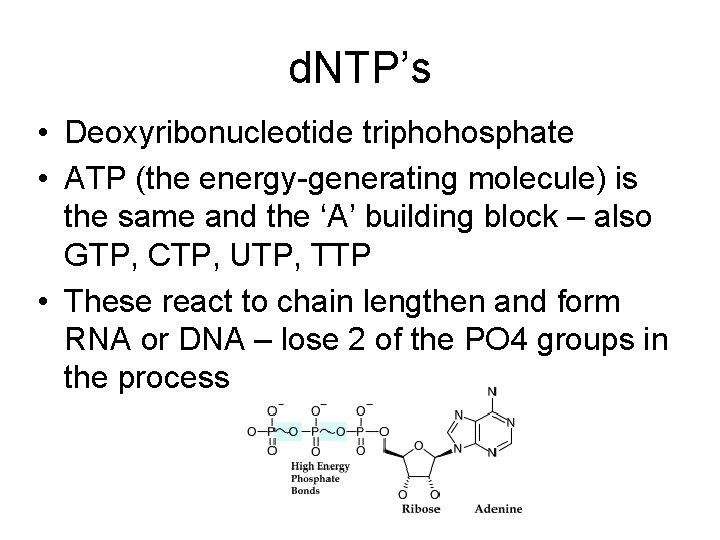
d. NTP’s • Deoxyribonucleotide triphohosphate • ATP (the energy-generating molecule) is the same and the ‘A’ building block – also GTP, CTP, UTP, TTP • These react to chain lengthen and form RNA or DNA – lose 2 of the PO 4 groups in the process

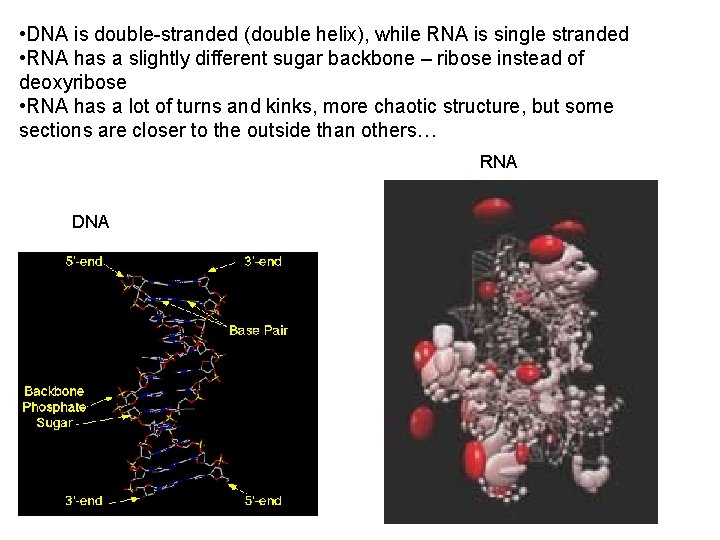
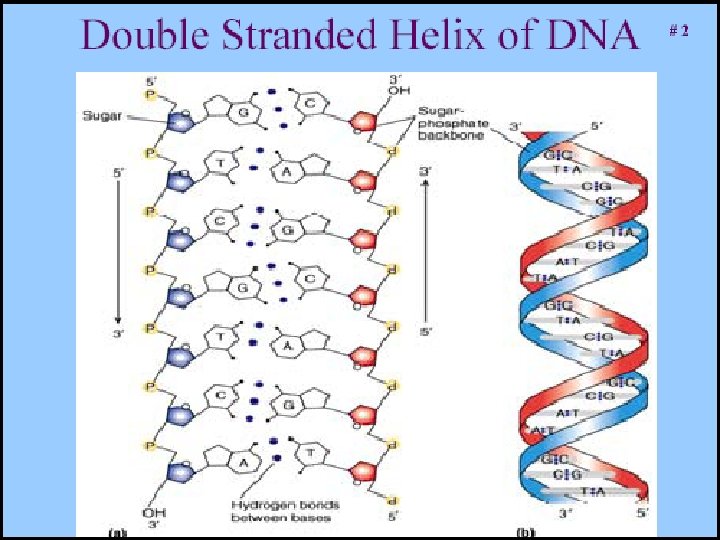
• DNA is double-stranded (double helix), while RNA is single stranded • RNA has a slightly different sugar backbone – ribose instead of deoxyribose • RNA has a lot of turns and kinks, more chaotic structure, but some sections are closer to the outside than others… RNA DNA


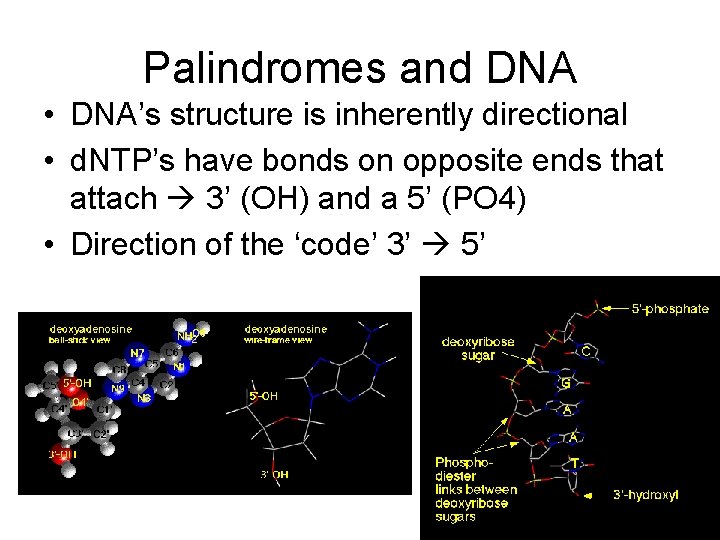
Palindromes and DNA • DNA’s structure is inherently directional • d. NTP’s have bonds on opposite ends that attach 3’ (OH) and a 5’ (PO 4) • Direction of the ‘code’ 3’ 5’


DNA enzymes • Restriction endonuclease – cuts DNA at specific base combinations • DNA ligase – links DNA molecules • DNA polymerase I – attaches DNTP’s, repairs DNA • DNA polymerase II – attaches DNTP’s, proofreads • DNA gyrase – twists, coils DNA • DNA Helicase – DNA strand separation • DNAse - degrades DNA to DNTP’s

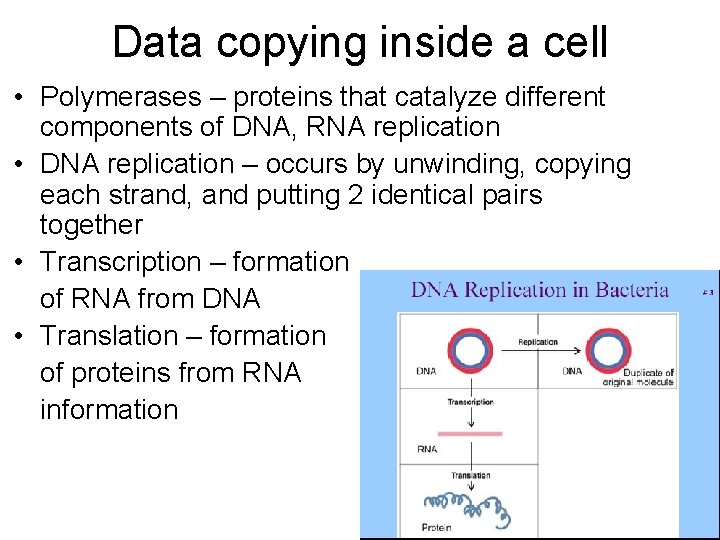
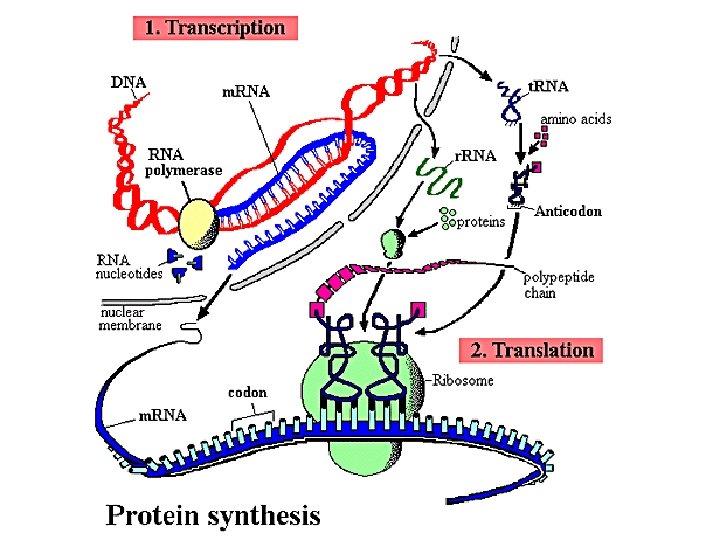
Data copying inside a cell • Polymerases – proteins that catalyze different components of DNA, RNA replication • DNA replication – occurs by unwinding, copying each strand, and putting 2 identical pairs together • Transcription – formation of RNA from DNA • Translation – formation of proteins from RNA information

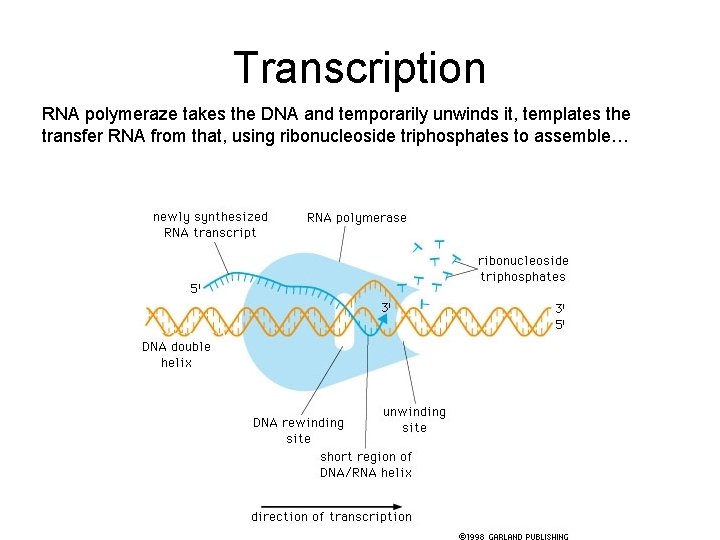
Transcription RNA polymeraze takes the DNA and temporarily unwinds it, templates the transfer RNA from that, using ribonucleoside triphosphates to assemble…

Ribosome • The ribosome is the site of translation of messenger RNA into protein. It is composed of two subunits. • In prokaryotes, the large subunit is called 50 S and the small subunit is called 30 S. The 30 S subunit consists of a single strand of RNA (the 16 S r. RNA, 1542 bases), and 21 proteins ranging in molecular weight from 9 k. D to 61 k. D. • The 30 S subunit is the site of translation initiation. • Measured by a ‘sedimentation coefficient’ – 16 S r. DNA is associated with a 16 S sized small subunit of the RNA translating ribosome

RNA and protein construction • The nucleotide base sequence of m. RNA is encoded from DNA and transmits sequences of bases used to determine the amino acid sequence of the protein. • m. RNA (“Messenger RNA”) associates with the ribosome (m. RNA and protein portion). • RNA (“Transfer RNA”) also required • Codons are 3 base m. RNA segments that specify a certain amino acid. • Most amino acids are coded for by more than one codon. • Translation ends when ribosome reached “stop codon” on m. RNA.

Ribosomal RNA • Ribosomal RNA is single stranded • RNA is a single stranded nucleic acid – m. RNA- messanger RNA – copies information from DNA and carries it to the ribosomes – t. RNA – transfers specific amino acids to the ribosomes – r. RNA – ribosomal RNA – with proteins, assembles ribosomal subunits DNA is transcribed to produce m. RNA then translated into proteins.

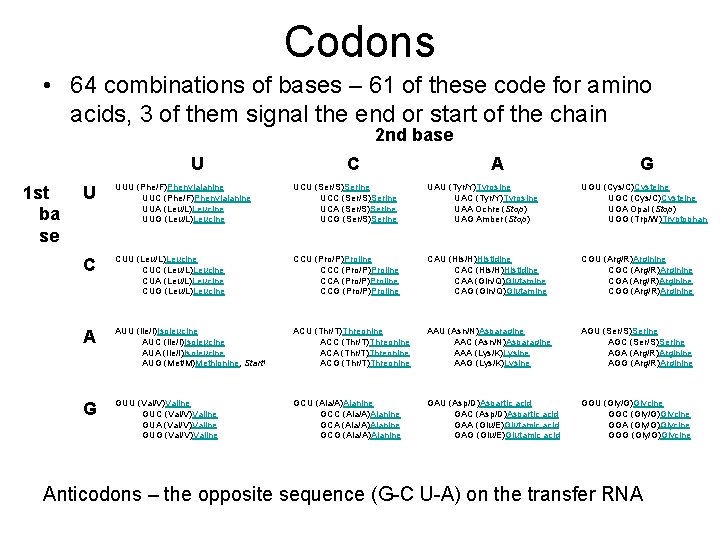
Codons • 64 combinations of bases – 61 of these code for amino acids, 3 of them signal the end or start of the chain 2 nd base U 1 st ba se C A G U UUU (Phe/F)Phenylalanine UUC (Phe/F)Phenylalanine UUA (Leu/L)Leucine UUG (Leu/L)Leucine UCU (Ser/S)Serine UCC (Ser/S)Serine UCA (Ser/S)Serine UCG (Ser/S)Serine UAU (Tyr/Y)Tyrosine UAC (Tyr/Y)Tyrosine UAA Ochre (Stop) UAG Amber (Stop) UGU (Cys/C)Cysteine UGC (Cys/C)Cysteine UGA Opal (Stop) UGG (Trp/W)Tryptophan C CUU (Leu/L)Leucine CUC (Leu/L)Leucine CUA (Leu/L)Leucine CUG (Leu/L)Leucine CCU (Pro/P)Proline CCC (Pro/P)Proline CCA (Pro/P)Proline CCG (Pro/P)Proline CAU (His/H)Histidine CAC (His/H)Histidine CAA (Gln/Q)Glutamine CAG (Gln/Q)Glutamine CGU (Arg/R)Arginine CGC (Arg/R)Arginine CGA (Arg/R)Arginine CGG (Arg/R)Arginine A AUU (Ile/I)Isoleucine AUC (Ile/I)Isoleucine AUA (Ile/I)Isoleucine AUG (Met/M)Methionine, Start 1 ACU (Thr/T)Threonine ACC (Thr/T)Threonine ACA (Thr/T)Threonine ACG (Thr/T)Threonine AAU (Asn/N)Asparagine AAC (Asn/N)Asparagine AAA (Lys/K)Lysine AAG (Lys/K)Lysine AGU (Ser/S)Serine AGC (Ser/S)Serine AGA (Arg/R)Arginine AGG (Arg/R)Arginine G GUU (Val/V)Valine GUC (Val/V)Valine GUA (Val/V)Valine GUG (Val/V)Valine GCU (Ala/A)Alanine GCC (Ala/A)Alanine GCA (Ala/A)Alanine GCG (Ala/A)Alanine GAU (Asp/D)Aspartic acid GAC (Asp/D)Aspartic acid GAA (Glu/E)Glutamic acid GAG (Glu/E)Glutamic acid GGU (Gly/G)Glycine GGC (Gly/G)Glycine GGA (Gly/G)Glycine GGG (Gly/G)Glycine Anticodons – the opposite sequence (G-C U-A) on the transfer RNA

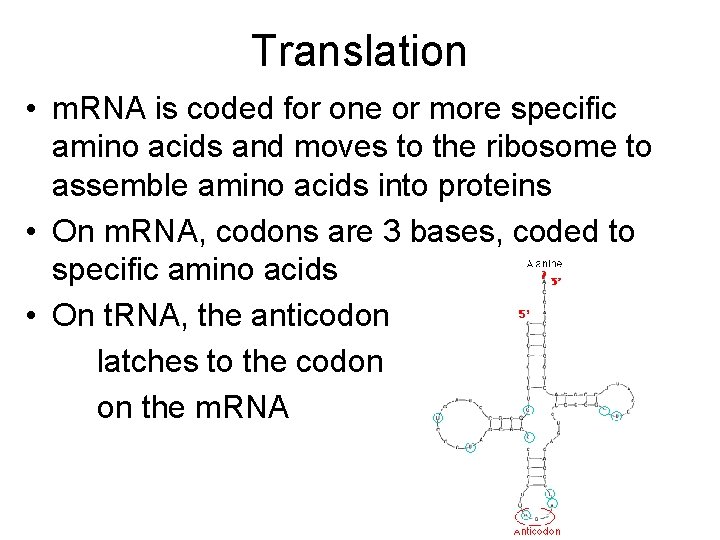
Translation • m. RNA is coded for one or more specific amino acids and moves to the ribosome to assemble amino acids into proteins • On m. RNA, codons are 3 bases, coded to specific amino acids • On t. RNA, the anticodon latches to the codon on the m. RNA

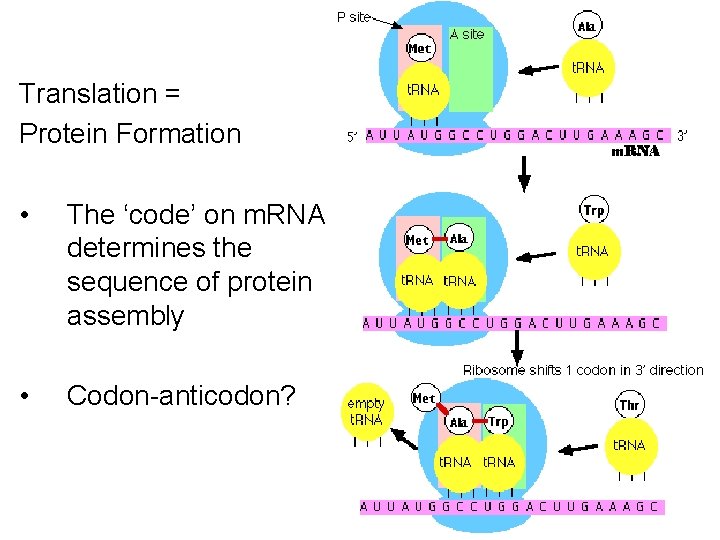
Translation = Protein Formation • The ‘code’ on m. RNA determines the sequence of protein assembly • Codon-anticodon?
