CDA Implementation Guide for Genetic Testing Report GTR





- Slides: 25

CDA Implementation Guide for Genetic Testing Report (GTR): Towards a Clinical Genomic Statement IHIC 2011 – Orlando, FL Amnon Shabo (Shvo), Ph. D shabo@il. ibm. com HL 7 Clinical Genomics WG Co-chair and Modeling Facilitator HL 7 Structured Documents WG CDA R 2 Co-editor CCD Implementation Guide Co-editor

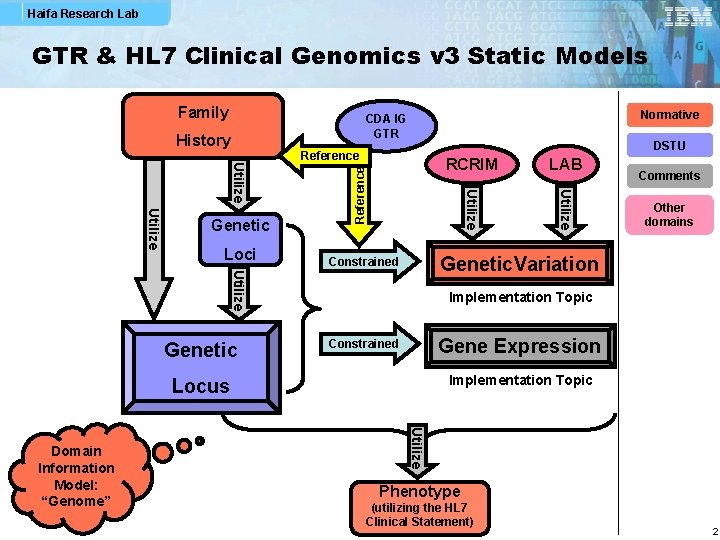
Haifa Research Lab GTR & HL 7 Clinical Genomics v 3 Static Models Family Normative CDA IG GTR Other domains Implementation Topic Gene Expression Constrained Implementation Topic Locus Utilize Domain Information Model: “Genome” Comments Genetic. Variation Constrained Utilize Genetic LAB Utilize Loci RCRIM Utilize Genetic DSTU Reference History Phenotype (utilizing the HL 7 Clinical Statement) 2

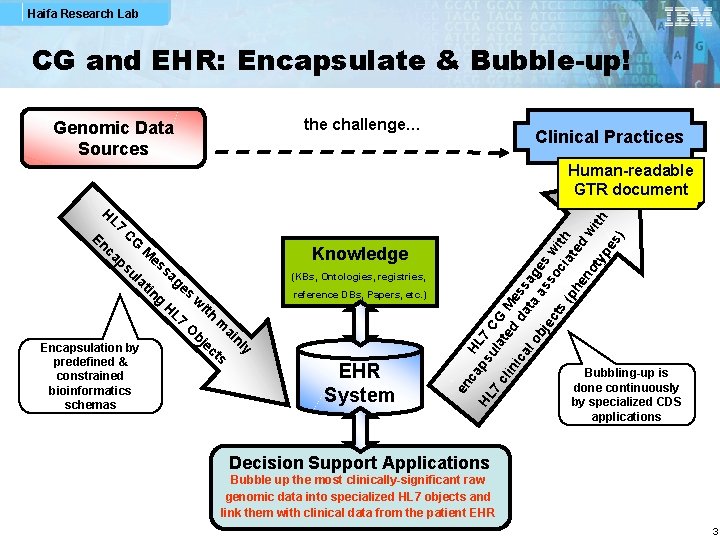
Haifa Research Lab CG and EHR: Encapsulate & Bubble-up! the challenge… Genomic Data Sources Clinical Practices predefined & constrained bioinformatics schemas Knowledge (KBs, Ontologies, registries, reference DBs, Papers, etc. ) EHR System HL En CG ca M es ps sa ul ge at in s g w H L 7 ith m O ai bj nl e y Encapsulation by ct s en c H L 7 ap HL su 7 C 7 la G cl te M in d ic da ess al ta ag ob as es je so w ct s ci ith (p at e he no d w ith ty pe s) Human-readable GTR document Bubbling-up is done continuously by specialized CDS applications Decision Support Applications Bubble up the most clinically-significant raw genomic data into specialized HL 7 objects and link them with clinical data from the patient EHR 3

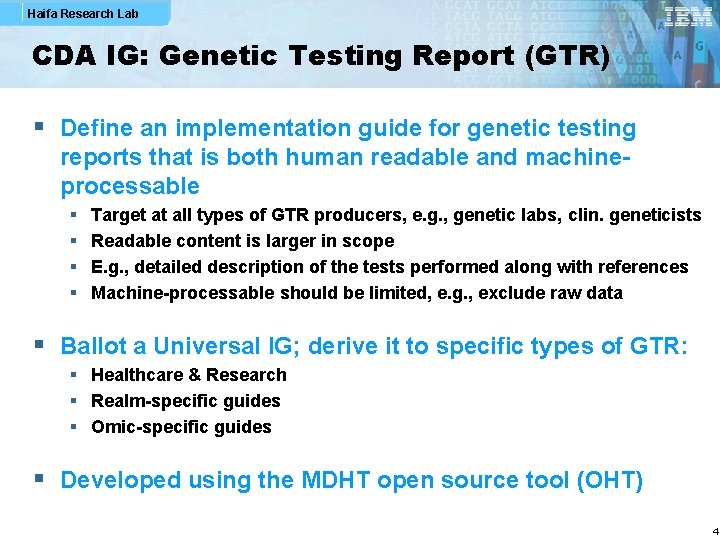
Haifa Research Lab CDA IG: Genetic Testing Report (GTR) § Define an implementation guide for genetic testing reports that is both human readable and machineprocessable § § Target at all types of GTR producers, e. g. , genetic labs, clin. geneticists Readable content is larger in scope E. g. , detailed description of the tests performed along with references Machine-processable should be limited, e. g. , exclude raw data § Ballot a Universal IG; derive it to specific types of GTR: § Healthcare & Research § Realm-specific guides § Omic-specific guides § Developed using the MDHT open source tool (OHT) 4

Haifa Research Lab GTR - Design Principles § Follow existing report formats commonly used in healthcare & research § Emphasize interpretations & recommendations § Provide general background information on tests performed § Reference HL 7 Clinical Genomics instances (e. g. , Genetic. Variation and Pedigree) as the place holders of full-blown raw genomic data and fully-structured family history data § Utilize patterns of ‘genotype-phenotype’ associations in the HL 7 v 3 Clinical Genomics Domain § Implement them as ‘clinical genomic statement’ entry-level templates (see next slide) 5

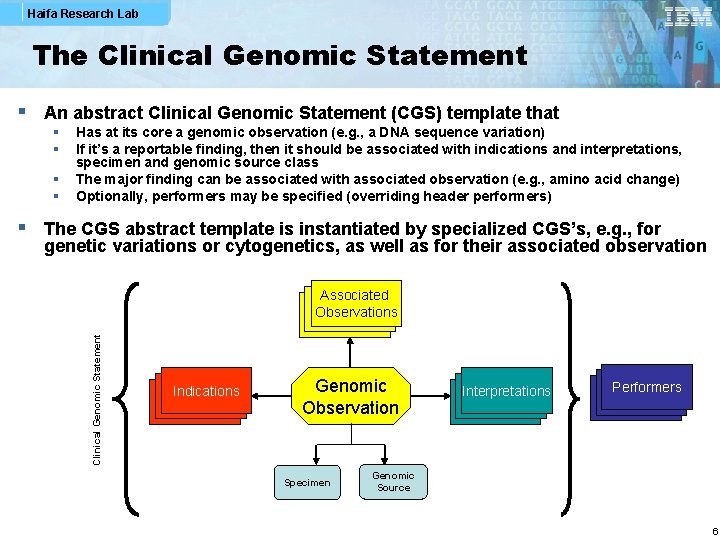
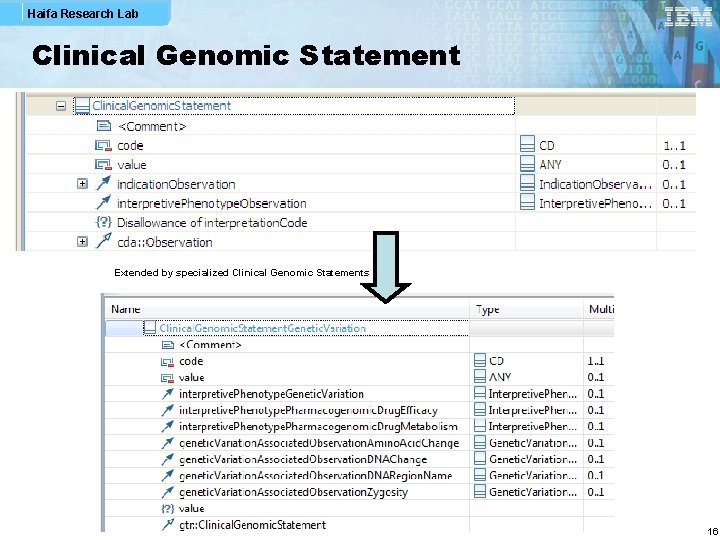
Haifa Research Lab The Clinical Genomic Statement § An abstract Clinical Genomic Statement (CGS) template that § § Has at its core a genomic observation (e. g. , a DNA sequence variation) If it’s a reportable finding, then it should be associated with indications and interpretations, specimen and genomic source class The major finding can be associated with associated observation (e. g. , amino acid change) Optionally, performers may be specified (overriding header performers) § The CGS abstract template is instantiated by specialized CGS’s, e. g. , for genetic variations or cytogenetics, as well as for their associated observation Clinical Genomic Statement Associated Observations Indications Genomic Observation Specimen Interpretations Performers Genomic Source 6

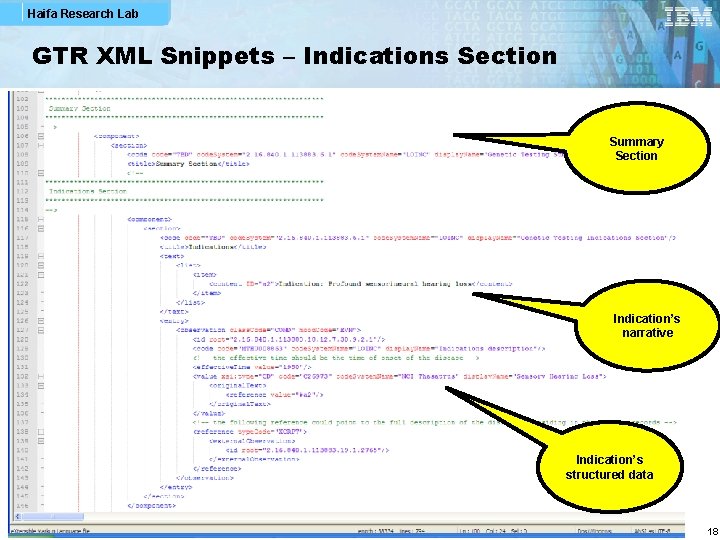
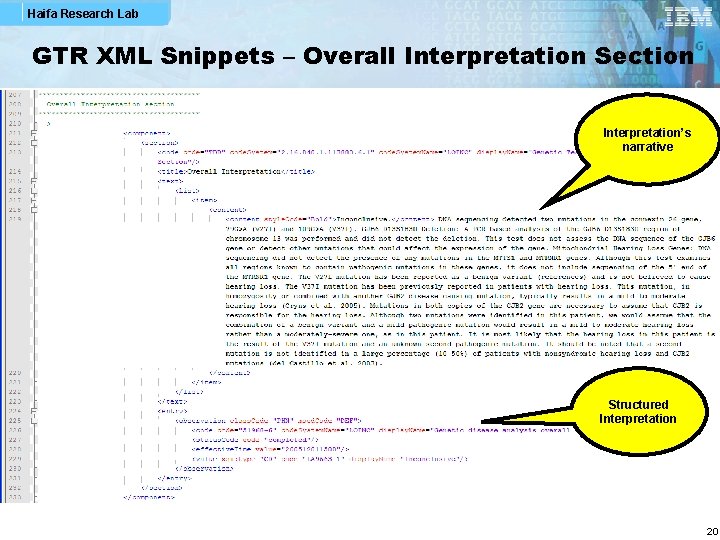
Haifa Research Lab Narrative and Structured Data § All CGS structured data items shall be part of clinical genomic statement (CGS) instances so that parsing applications can find the full semantics explicitly represented in one coherent structure § Sub-sections such as Indications, Interpretations and Specimen are mainly for presenting narrative, but they may also contain structured data § In this way, it is possible to have less redundant documents, e. g. , in the case where all tests reported in a GTR document have the same indication, an Indications section in the Summary section consists of a full-blown indication observation which all CGS indication observations reference § CGS structured data may point to the respective narrative in subsections (by means of XML ID) 7

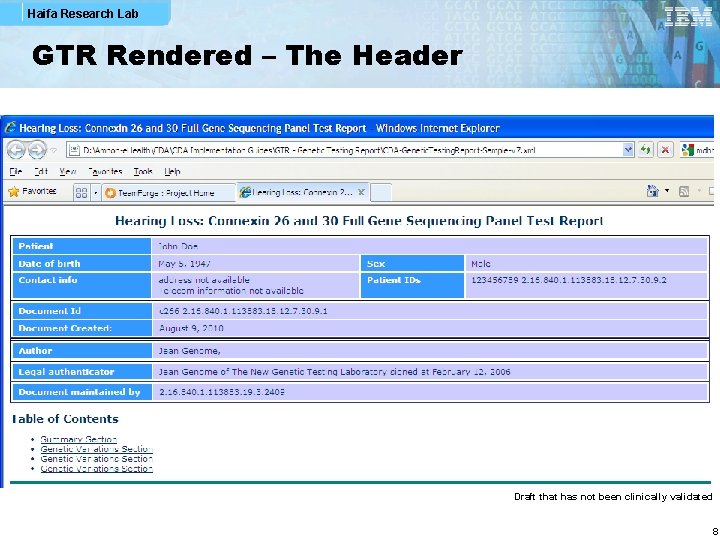
Haifa Research Lab GTR Rendered – The Header Draft that has not been clinically validated 8

Haifa Research Lab GTR Rendered – Summary Section Draft that has not been clinically validated Summary Section continues in next slide… 9

Haifa Research Lab GTR Rendered – Summary Section (cont. ) Draft that has not been clinically validated 10

Haifa Research Lab GTR Rendered – Genetic Variation Sections Draft that has not been clinically validated 11

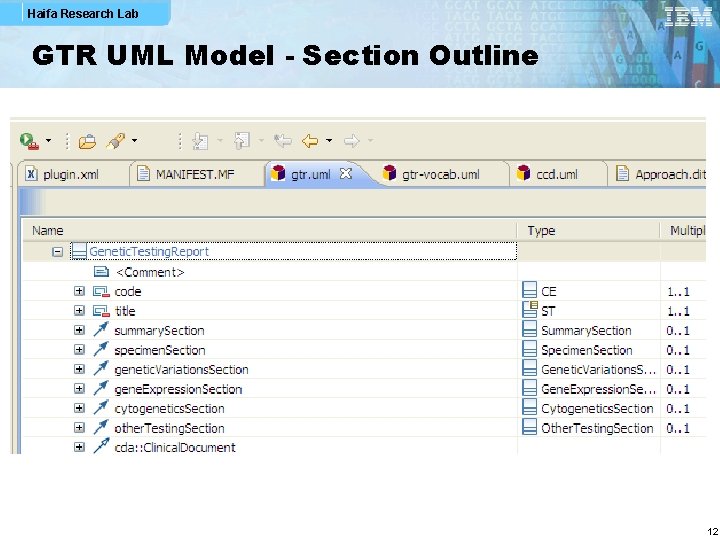
Haifa Research Lab GTR UML Model - Section Outline 12

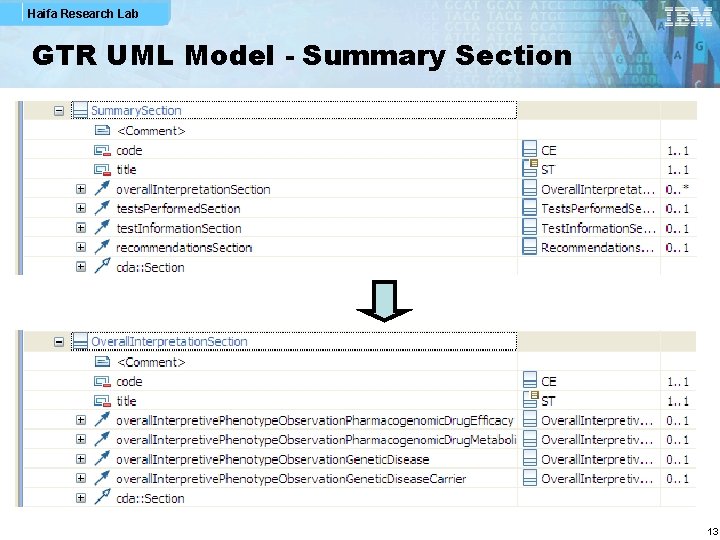
Haifa Research Lab GTR UML Model - Summary Section 13

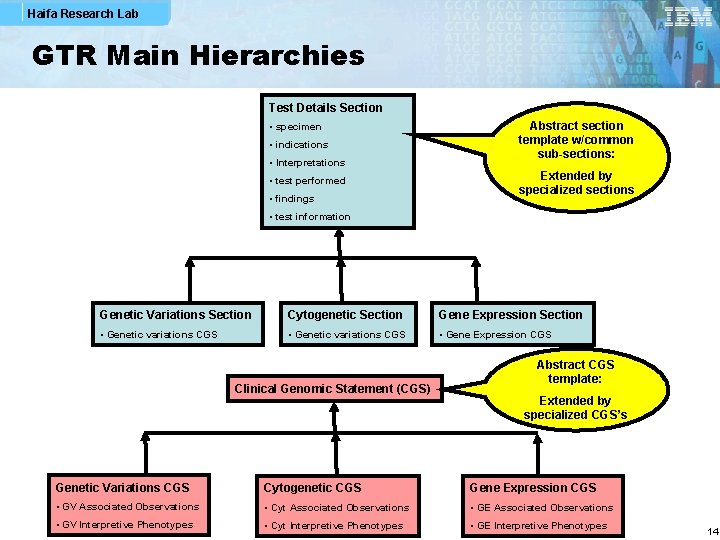
Haifa Research Lab GTR Main Hierarchies Test Details Section • specimen • indications • Interpretations • test performed • findings Abstract section template w/common sub-sections: Extended by specialized sections • test information Genetic Variations Section Cytogenetic Section Gene Expression Section • Genetic variations CGS • Gene Expression CGS Clinical Genomic Statement (CGS) Abstract CGS template: Extended by specialized CGS’s Genetic Variations CGS Cytogenetic CGS Gene Expression CGS • GV Associated Observations • Cyt Associated Observations • GE Associated Observations • GV Interpretive Phenotypes • Cyt Interpretive Phenotypes • GE Interpretive Phenotypes 14

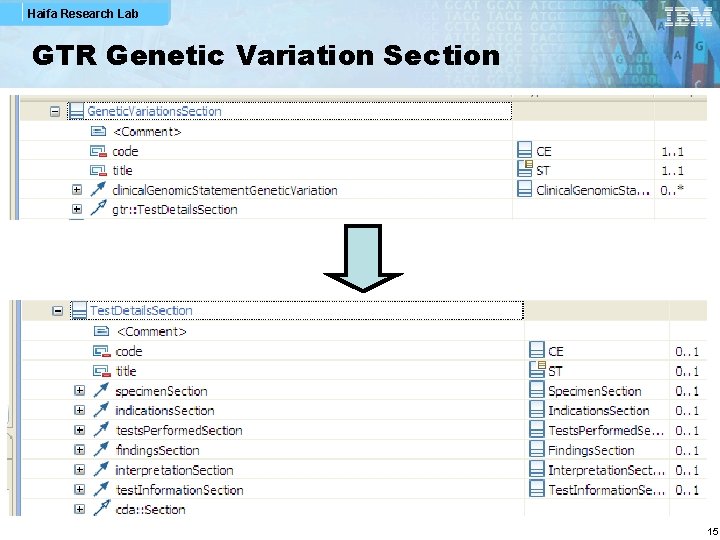
Haifa Research Lab GTR Genetic Variation Section 15

Haifa Research Lab Clinical Genomic Statement Extended by specialized Clinical Genomic Statements 16

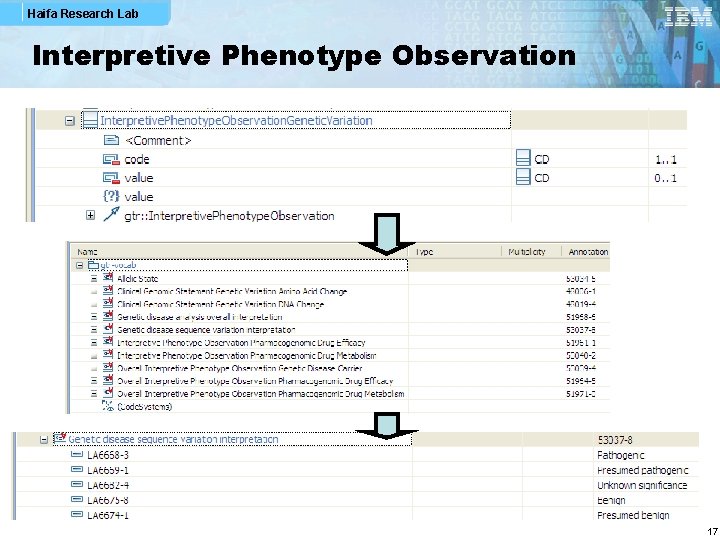
Haifa Research Lab Interpretive Phenotype Observation 17

Haifa Research Lab GTR XML Snippets – Indications Section Summary Section Indication’s narrative Indication’s structured data 18

Haifa Research Lab GTR XML Snippets – Specimen Section Specimen’s narrative Specimen’s structured data 19

Haifa Research Lab GTR XML Snippets – Overall Interpretation Section Interpretation’s narrative Structured Interpretation 20

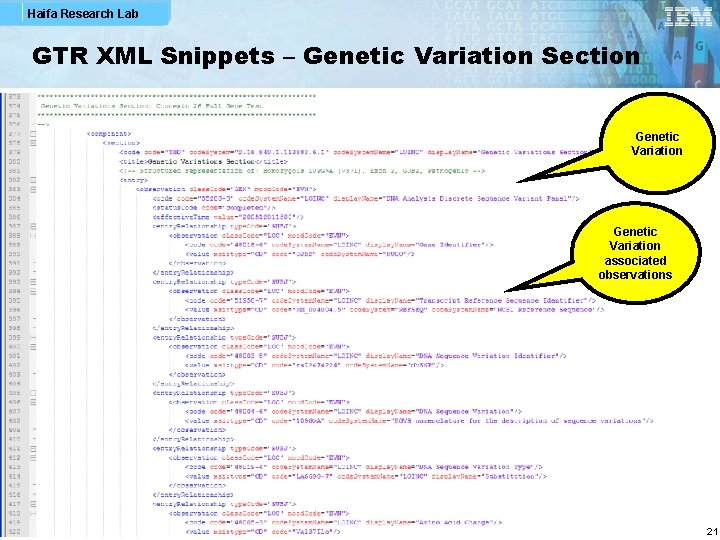
Haifa Research Lab GTR XML Snippets – Genetic Variation Section Genetic Variation associated observations 21

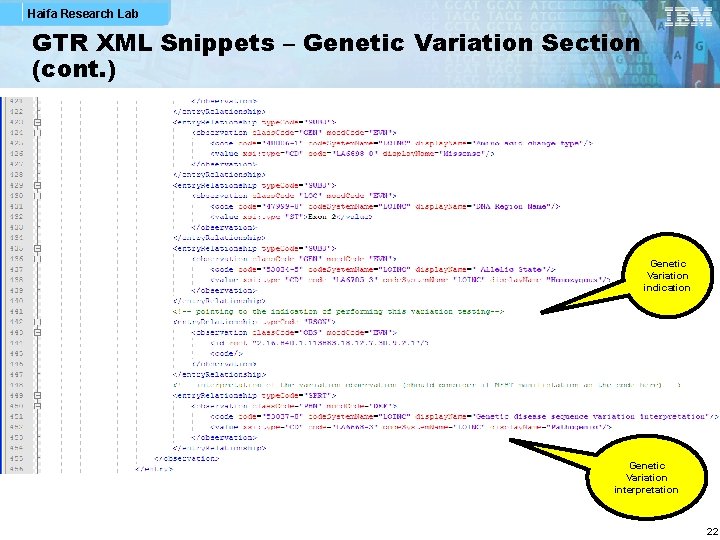
Haifa Research Lab GTR XML Snippets – Genetic Variation Section (cont. ) Genetic Variation indication Genetic Variation interpretation 22

Haifa Research Lab u. Health: Patient Empowerment System § Joint project between IBM and Gil Hospital )Korea( § 5 years project, currently in 2 nd year § 3 IBM centers are involved: § IBM Korea § IBM Research in China § IBM Research in Haifa § Building an open platform and services § Web-based rich portal § Testing the system with patients and physicians from the hospital § Support integration with external PHR (e. g. Google Health) 23

Haifa Research Lab u. Health: PHR / EHR Hybrid System Using CCD + GTR 24

Haifa Research Lab The End • Thank you for your attention… • Questions? 25